Context
I am the author and maintainer of netgraph, a python library for creating network visualisations.
I am currently trying to optimise a routine that computes a set of N node positions for networks in which each edge has a defined length.
An example can be found here.
Problem
At its core, the routine runs scipy.optimize.minimize to compute the positions that maximise the total distance between nodes:
def cost_function(positions):
return 1. / np.sum((pdist(positions.reshape((-1, 2))))**power)
result = minimize(cost_function, initial_positions.flatten(), method='SLSQP',
jac="2-point", constraints=[nonlinear_constraint])
positionsare an (unravelled) numpy array of (x, y) tuples.poweris a smallish number that limits the influence of large distances (to encourage compact node layouts) but for the purpose of this question could be assumed to be 1.pdistis the pairwise distance function inscipy.spatial.
The minimisation ( / maximisation) is constrained using the following non-linear constraint:
lower_bounds = ... # (squareform of an) (N, N) distance matrix of the sum of node sizes (i.e. nodes should not overlap)
upper_bounds = ... # (squareform of an) (N, N) distance matrix constructed from the given edge lengths
def constraint_function(positions):
positions = np.reshape(positions, (-1, 2))
return pdist(positions)
nonlinear_constraint = NonlinearConstraint(constraint_function, lb=lower_bounds, ub=upper_bounds, jac='2-point')
For toy examples, the optimisation completes correctly and quickly. However, even for smallish networks, the running time is fairly abysmal.
My current implementation uses finite differences to approximate the gradients (jac='2-point').
To speed up the computation, I would like to compute the Jacobians explicitly.
Following several Math Stackexchange posts (1, 2), I computed the the Jacobian of the pairwise distance function as follows:
def delta_constraint(positions):
positions = np.reshape(positions, (-1, 2))
total_positions = positions.shape[0]
delta = positions[np.newaxis, :, :] - positions[:, np.newaxis, :]
distance = np.sqrt(np.sum(delta ** 2, axis=-1))
jac = delta / distance[:, :, np.newaxis]
squareform_indices = np.triu_indices(total_positions, 1)
return jac[squareform_indices]
nonlinear_constraint = NonlinearConstraint(constraint_function, lb=lower_bounds, ub=upper_bounds, jac=delta_constraint)
However, this results in a ValueError, as the shape of the output is incorrect. For the triangle example, the expected output shape is (3, 6), whereas the function above returns a (3, 2) array (i.e. 3 pairwise distance by 2 dimensions). For the square the expected output is (6, 8), whereas the actual is (6, 2). Any help deriving implementing the correct callable(s) for the jac arguments to NonlinearConstraint and minimize would be appreciated.
Note
I would like to avoid the use of autograd/jax/numdifftools (as in this question), as I would like to keep the number of dependencies of my library small.
Minimal working example(s)
#!/usr/bin/env python
"""
Create a node layout with fixed edge lengths but unknown node positions.
"""
import numpy as np
from scipy.optimize import minimize, NonlinearConstraint
from scipy.spatial.distance import pdist, squareform
def get_geometric_node_layout(edges, edge_length, node_size=0., power=0.2, maximum_iterations=200, origin=(0, 0), scale=(1, 1)):
"""Node layout for defined edge lengths but unknown node positions.
Node positions are determined through non-linear optimisation: the
total distance between nodes is maximised subject to the constraint
imposed by the edge lengths, which are used as upper bounds.
If provided, node sizes are used to set lower bounds.
Parameters
----------
edges : list
The edges of the graph, with each edge being represented by a (source node ID, target node ID) tuple.
edge_lengths : dict
Mapping of edges to their lengths.
node_size : scalar or dict, default 0.
Size (radius) of nodes.
Providing the correct node size minimises the overlap of nodes in the graph,
which can otherwise occur if there are many nodes, or if the nodes differ considerably in size.
power : float, default 0.2.
The cost being minimised is the inverse of the sum of distances.
The power parameter is the exponent applied to each distance before summation.
Large values result in positions that are stretched along one axis.
Small values decrease the influence of long distances on the cost
and promote a more compact layout.
maximum_iterations : int
Maximum number of iterations of the minimisation.
origin : tuple, default (0, 0)
The (float x, float y) coordinates corresponding to the lower left hand corner of the bounding box specifying the extent of the canvas.
scale : tuple, default (1, 1)
The (float x, float y) dimensions representing the width and height of the bounding box specifying the extent of the canvas.
Returns
-------
node_positions : dict
Dictionary mapping each node ID to (float x, float y) tuple, the node position.
"""
# TODO: assert triangle inequality
# TODO: assert that the edges fit within the canvas dimensions
# ensure that graph is bi-directional
edges = edges + [(target, source) for (source, target) in edges] # forces copy
edges = list(set(edges))
# upper bound: pairwise distance matrix with unknown distances set to the maximum possible distance given the canvas dimensions
lengths = []
for (source, target) in edges:
if (source, target) in edge_length:
lengths.append(edge_length[(source, target)])
else:
lengths.append(edge_length[(target, source)])
sources, targets = zip(*edges)
nodes = sources + targets
unique_nodes = set(nodes)
indices = range(len(unique_nodes))
node_to_idx = dict(zip(unique_nodes, indices))
source_indices = [node_to_idx[source] for source in sources]
target_indices = [node_to_idx[target] for target in targets]
total_nodes = len(unique_nodes)
max_distance = np.sqrt(scale[0]**2 + scale[1]**2)
distance_matrix = np.full((total_nodes, total_nodes), max_distance)
distance_matrix[source_indices, target_indices] = lengths
distance_matrix[np.diag_indices(total_nodes)] = 0
upper_bounds = squareform(distance_matrix)
# lower bound: sum of node sizes
if isinstance(node_size, (int, float)):
sizes = node_size * np.ones((total_nodes))
elif isinstance(node_size, dict):
sizes = np.array([node_size[node] if node in node_size else 0. for node in unique_nodes])
sum_of_node_sizes = sizes[np.newaxis, :] + sizes[:, np.newaxis]
sum_of_node_sizes -= np.diag(np.diag(sum_of_node_sizes)) # squareform requires zeros on diagonal
lower_bounds = squareform(sum_of_node_sizes)
def cost_function(positions):
return 1. / np.sum((pdist(positions.reshape((-1, 2))))**power)
def constraint_function(positions):
positions = np.reshape(positions, (-1, 2))
return pdist(positions)
initial_positions = _initialise_geometric_node_layout(edges)
nonlinear_constraint = NonlinearConstraint(constraint_function, lb=lower_bounds, ub=upper_bounds, jac='2-point')
result = minimize(cost_function, initial_positions.flatten(), method='SLSQP',
jac="2-point", constraints=[nonlinear_constraint], options=dict(maxiter=maximum_iterations))
if not result.success:
print("Warning: could not compute valid node positions for the given edge lengths.")
print(f"scipy.optimize.minimize: {result.message}.")
node_positions_as_array = result.x.reshape((-1, 2))
node_positions = dict(zip(unique_nodes, node_positions_as_array))
return node_positions
def _initialise_geometric_node_layout(edges):
sources, targets = zip(*edges)
total_nodes = len(set(sources + targets))
return np.random.rand(total_nodes, 2)
if __name__ == '__main__':
import matplotlib.pyplot as plt
def plot_graph(edges, node_layout):
# poor man's graph plotting
fig, ax = plt.subplots()
for source, target in edges:
x1, y1 = node_layout[source]
x2, y2 = node_layout[target]
ax.plot([x1, x2], [y1, y2], color='darkgray')
ax.set_aspect('equal')
################################################################################
# triangle with right angle
edges = [
(0, 1),
(1, 2),
(2, 0)
]
lengths = {
(0, 1) : 3,
(1, 2) : 4,
(2, 0) : 5,
}
pos = get_geometric_node_layout(edges, lengths, node_size=0)
plot_graph(edges, node_layout=pos)
plt.show()
################################################################################
# square
edges = [
(0, 1),
(1, 2),
(2, 3),
(3, 0),
]
lengths = {
(0, 1) : 0.5,
(1, 2) : 0.5,
(2, 3) : 0.5,
(3, 0) : 0.5,
}
pos = get_geometric_node_layout(edges, lengths, node_size=0)
plot_graph(edges, node_layout=pos)
plt.show()
Edit: Realistic use case for timing
Below is a more realistic use case that I am using to time my code.
I have incorporated @adrianop01's computation of the Jacobian for the constraint. It also includes a superior initialisation. It requires the additional dependencies networkx and netgraph, both of which can be installed via pip.
#!/usr/bin/env python
"""
Create a node layout with fixed edge lengths but unknown node positions.
"""
import numpy as np
from itertools import combinations
from scipy.optimize import minimize, NonlinearConstraint
from scipy.spatial.distance import pdist, squareform
from netgraph._node_layout import _rescale_to_frame
def get_geometric_node_layout(edges, edge_length, node_size=0., power=0.2, maximum_iterations=200, origin=(0, 0), scale=(1, 1)):
"""Node layout for defined edge lengths but unknown node positions.
Node positions are determined through non-linear optimisation: the
total distance between nodes is maximised subject to the constraint
imposed by the edge lengths, which are used as upper bounds.
If provided, node sizes are used to set lower bounds.
Parameters
----------
edges : list
The edges of the graph, with each edge being represented by a (source node ID, target node ID) tuple.
edge_lengths : dict
Mapping of edges to their lengths.
node_size : scalar or dict, default 0.
Size (radius) of nodes.
Providing the correct node size minimises the overlap of nodes in the graph,
which can otherwise occur if there are many nodes, or if the nodes differ considerably in size.
power : float, default 0.2.
The cost being minimised is the inverse of the sum of distances.
The power parameter is the exponent applied to each distance before summation.
Large values result in positions that are stretched along one axis.
Small values decrease the influence of long distances on the cost
and promote a more compact layout.
maximum_iterations : int
Maximum number of iterations of the minimisation.
origin : tuple, default (0, 0)
The (float x, float y) coordinates corresponding to the lower left hand corner of the bounding box specifying the extent of the canvas.
scale : tuple, default (1, 1)
The (float x, float y) dimensions representing the width and height of the bounding box specifying the extent of the canvas.
Returns
-------
node_positions : dict
Dictionary mapping each node ID to (float x, float y) tuple, the node position.
"""
# TODO: assert triangle inequality
# TODO: assert that the edges fit within the canvas dimensions
# ensure that graph is bi-directional
edges = edges + [(target, source) for (source, target) in edges] # forces copy
edges = list(set(edges))
# upper bound: pairwise distance matrix with unknown distances set to the maximum possible distance given the canvas dimensions
lengths = []
for (source, target) in edges:
if (source, target) in edge_length:
lengths.append(edge_length[(source, target)])
else:
lengths.append(edge_length[(target, source)])
sources, targets = zip(*edges)
nodes = sources + targets
unique_nodes = set(nodes)
indices = range(len(unique_nodes))
node_to_idx = dict(zip(unique_nodes, indices))
source_indices = [node_to_idx[source] for source in sources]
target_indices = [node_to_idx[target] for target in targets]
total_nodes = len(unique_nodes)
max_distance = np.sqrt(scale[0]**2 + scale[1]**2)
distance_matrix = np.full((total_nodes, total_nodes), max_distance)
distance_matrix[source_indices, target_indices] = lengths
distance_matrix[np.diag_indices(total_nodes)] = 0
upper_bounds = squareform(distance_matrix)
# lower bound: sum of node sizes
if isinstance(node_size, (int, float)):
sizes = node_size * np.ones((total_nodes))
elif isinstance(node_size, dict):
sizes = np.array([node_size[node] if node in node_size else 0. for node in unique_nodes])
sum_of_node_sizes = sizes[np.newaxis, :] + sizes[:, np.newaxis]
sum_of_node_sizes -= np.diag(np.diag(sum_of_node_sizes)) # squareform requires zeros on diagonal
lower_bounds = squareform(sum_of_node_sizes)
invalid = lower_bounds > upper_bounds
lower_bounds[invalid] = upper_bounds[invalid] - 1e-8
def cost_function(positions):
# return -np.sum((pdist(positions.reshape((-1, 2))))**power)
return 1. / np.sum((pdist(positions.reshape((-1, 2))))**power)
def cost_jacobian(positions):
# TODO
pass
def constraint_function(positions):
positions = np.reshape(positions, (-1, 2))
return pdist(positions)
# adapted from https://mcmap.net/q/1342336/-how-to-compute-the-jacobian-of-a-pairwise-distance-function-scipy-spatial-pdist
total_pairs = int((total_nodes - 1) * total_nodes / 2)
source_indices, target_indices = np.array(list(combinations(range(total_nodes), 2))).T # node order thus (0,1) ... (0,N-1), (1,2),...(1,N-1),...,(N-2,N-1)
rows = np.repeat(np.arange(total_pairs).reshape(-1, 1), 2, axis=1)
source_columns = np.vstack((source_indices*2, source_indices*2+1)).T
target_columns = np.vstack((target_indices*2, target_indices*2+1)).T
def constraint_jacobian(positions):
positions = np.reshape(positions, (-1, 2))
pairwise_distances = constraint_function(positions)
jac = np.zeros((total_pairs, 2 * total_nodes))
jac[rows, source_columns] = (positions[source_indices] - positions[target_indices]) / pairwise_distances.reshape((-1, 1))
jac[rows, target_columns] = -jac[rows, source_columns]
return jac
initial_positions = _initialise_geometric_node_layout(edges, edge_length)
nonlinear_constraint = NonlinearConstraint(constraint_function, lb=lower_bounds, ub=upper_bounds, jac=constraint_jacobian)
result = minimize(cost_function, initial_positions.flatten(), method='SLSQP',
jac='2-point', constraints=[nonlinear_constraint], options=dict(maxiter=maximum_iterations))
# result = minimize(cost_function, initial_positions.flatten(), method='trust-constr',
# jac=cost_jacobian, constraints=[nonlinear_constraint])
if not result.success:
print("Warning: could not compute valid node positions for the given edge lengths.")
print(f"scipy.optimize.minimize: {result.message}.")
node_positions_as_array = result.x.reshape((-1, 2))
node_positions_as_array = _rescale_to_frame(node_positions_as_array, np.array(origin), np.array(scale))
node_positions = dict(zip(unique_nodes, node_positions_as_array))
return node_positions
# # slow
# def _initialise_geometric_node_layout(edges, edge_length=None):
# sources, targets = zip(*edges)
# total_nodes = len(set(sources + targets))
# return np.random.rand(total_nodes, 2)
# much faster
def _initialise_geometric_node_layout(edges, edge_length=None):
"""Initialises the node positions using the FR algorithm with weights.
Shorter edges are given a larger weight such that the nodes experience a strong attractive force."""
from netgraph import get_fruchterman_reingold_layout
if edge_length:
edge_weight = dict()
for edge, length in edge_length.items():
edge_weight[edge] = 1 / length
else:
edge_weight = None
node_positions = get_fruchterman_reingold_layout(edges)
return np.array(list(node_positions.values()))
if __name__ == '__main__':
from time import time
import matplotlib.pyplot as plt
import networkx as nx # pip install networkx
from netgraph import Graph # pip install netgraph
fig, (ax1, ax2) = plt.subplots(1, 2)
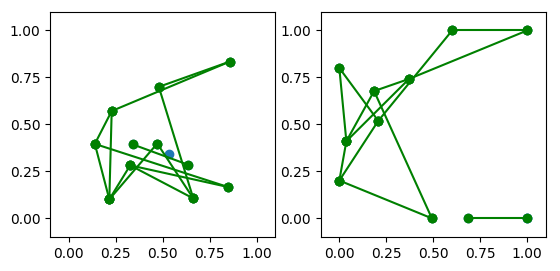
g = nx.random_geometric_graph(50, 0.3, seed=2)
node_positions = nx.get_node_attributes(g, 'pos')
plot_instance = Graph(g,
node_layout=node_positions,
node_size=1, # netgraph rescales node sizes by 0.01
node_edge_width=0.1,
edge_width=0.1,
ax=ax1,
)
ax1.axis([0, 1, 0, 1])
ax1.set_title('Original node positions')
def get_euclidean_distance(p1, p2):
return np.sqrt(np.sum((np.array(p1)-np.array(p2))**2))
edge_length = dict()
for (source, target) in g.edges:
edge_length[(source, target)] = get_euclidean_distance(node_positions[source], node_positions[target])
tic = time()
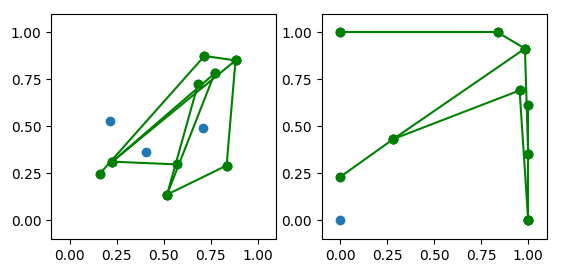
new_node_positions = get_geometric_node_layout(list(g.edges), edge_length, node_size=0.01)
toc = time()
print(f"Time elapsed : {toc-tic}")
Graph(g,
node_layout=new_node_positions,
node_size=1,
node_edge_width=0.1,
edge_width=0.1,
ax=ax2,
)
ax2.axis([0, 1, 0, 1])
ax2.set_title('Reconstructed node positions')
plt.show()
2nd edit
Here are some preliminary results I obtained when testing @spinkus' and related solutions. My implementation of his code looks like this:
def cost_function(positions):
return -np.sum((pdist(positions.reshape((-1, 2))))**2)
def cost_jacobian(positions):
positions = positions.reshape(-1, 2)
delta = positions[np.newaxis, :] - positions[:, np.newaxis]
jac = -2 * np.sum(delta, axis=0)
return jac.ravel()
Unfortunately, this cost function takes significantly longer to converge: 13 seconds in a best of 5 with a large variance in the timings (up to a minute). This is independent of whether I use the explicit Jacobian or approximate it using the finite difference approach. Furthermore, the minimisation often ends prematurely with "scipy.optimize.minimize: Inequality constraints incompatible." and "scipy.optimize.minimize: Positive directional derivative for linesearch." My bet (although I have little evidence to back it up) is that absolute value of the cost matters. My original cost function decreases both in value and in absolute value, whereas the minimisation increases the absolute value of the @spinkus cost function (however, see @spinkus' excellent comment below why that might be somewhat a red herring and lead to less accurate solutions).
I also understood (I think) why my original cost function is not amenable to computing the Jacobians.
Let power be 0.5, then the cost function and Jacobian take this form (unless my algebra is wrong again):
def cost_function(positions):
return 1. / np.sum((pdist(positions.reshape((-1, 2))))**0.5)
def cost_jacobian(positions):
positions = positions.reshape(-1, 2)
delta = positions[np.newaxis, :] - positions[:, np.newaxis]
distance = np.sqrt(np.sum(delta**2, axis=-1))
denominator = -2 * np.sqrt(delta) * distance[:, :, np.newaxis]
denominator[np.diag_indices_from(denominator[:, :, 0]),:] = 1
jac = 1 / denominator
return np.sum(jac, axis=0).ravel() - 1
The problematic term is the sqrt(delta), where delta are the vectors between all points.
Ignoring the diagonals, half of the entries in this matrix are necessarily negative and thus the Jacobian cannot be computed.
However, the purpose of the power is simply to decrease the importance of large distances on the cost.
Any monotonically increasing function with decreasing derivative will do.
Using log(x + 1) instead of the power results in these functions:
def cost_function(positions):
return 1 / np.sum(np.log(pdist(positions.reshape((-1, 2))) + 1))
def cost_jacobian(positions):
positions = positions.reshape(-1, 2)
delta = positions[np.newaxis, :] - positions[:, np.newaxis]
distance2 = np.sum(delta**2, axis=-1)
distance2[np.diag_indices_from(distance2)] = 1
jac = -delta / (distance2 + np.sqrt(distance2))[..., np.newaxis]
return np.sum(jac, axis=0).ravel()
With the finite difference approximation, the minimisation terminates in 0.5 seconds. However, with the explicit Jacobian, the best run times were 4 seconds albeit still with a very large variance with run times up a minute and longer.
Tl;dr.
I still don't understand why the minimisation does not run faster with explicit Jacobians.




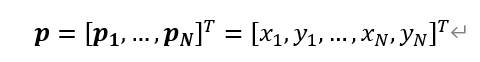

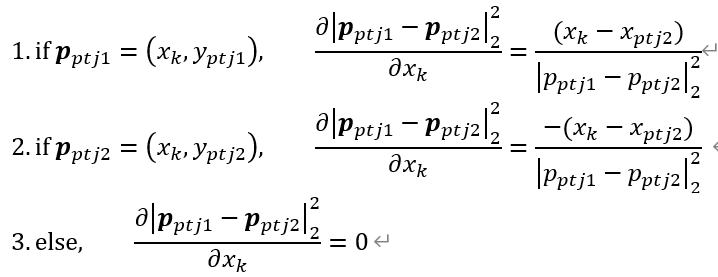

lower_boundsin OP's code – Unbuttonscaleparameter. – Graffpowerto 2 and changing the form of the objective from-1/xto-1x. Latter won't effect optimal. – Katar