I try to solve a common problem in medicine: the combination of a prediction model with other sources, eg, an expert opinion [sometimes heavily emphysised in medicine], called superdoc predictor in this post.
This could be solved by stack a model with a logistic regression (that enters the expert opinion) as described on page 26 in this paper:
Afshar P, Mohammadi A, Plataniotis KN, Oikonomou A, Benali H. From Handcrafted to Deep-Learning-Based Cancer Radiomics: Challenges and Opportunities. IEEE Signal Process Mag 2019; 36: 132–60. Available here
I've tried this here without considering overfitting (I did not apply out of fold predictions of the lower learner):
Example data
# library
library(tidyverse)
library(caret)
library(glmnet)
library(mlbench)
# get example data
data(PimaIndiansDiabetes, package="mlbench")
data <- PimaIndiansDiabetes
# add the super doctors opinion to the data
set.seed(2323)
data %>%
rowwise() %>%
mutate(superdoc=case_when(diabetes=="pos" ~ as.numeric(sample(0:2,1)), TRUE~ 0)) -> data
# separate the data in a training set and test set
train.data <- data[1:550,]
test.data <- data[551:768,]
Stacked models without considering out of fold predictions:
# elastic net regression (without the superdoc's opinion)
set.seed(2323)
model <- train(
diabetes ~., data = train.data %>% select(-superdoc), method = "glmnet",
trControl = trainControl("repeatedcv",
number = 10,
repeats=10,
classProbs = TRUE,
savePredictions = TRUE,
summaryFunction = twoClassSummary),
tuneLength = 10,
metric="ROC" #ROC metric is in twoClassSummary
)
# extract the coefficients for the best alpha and lambda
coef(model$finalModel, model$finalModel$lambdaOpt) -> coeffs
tidy(coeffs) %>% tibble() -> coeffs
coef.interc = coeffs %>% filter(row=="(Intercept)") %>% pull(value)
coef.pregnant = coeffs %>% filter(row=="pregnant") %>% pull(value)
coef.glucose = coeffs %>% filter(row=="glucose") %>% pull(value)
coef.pressure = coeffs %>% filter(row=="pressure") %>% pull(value)
coef.mass = coeffs %>% filter(row=="mass") %>% pull(value)
coef.pedigree = coeffs %>% filter(row=="pedigree") %>% pull(value)
coef.age = coeffs %>% filter(row=="age") %>% pull(value)
# combine the model with the superdoc's opinion in a logistic regression model
finalmodel = glm(diabetes ~ superdoc + I(coef.interc + coef.pregnant*pregnant + coef.glucose*glucose + coef.pressure*pressure + coef.mass*mass + coef.pedigree*pedigree + coef.age*age),family=binomial, data=train.data)
# make predictions on the test data
predict(finalmodel,test.data, type="response") -> predictions
# check the AUC of the model in the test data
roc(test.data$diabetes,predictions, ci=TRUE)
#> Setting levels: control = neg, case = pos
#> Setting direction: controls < cases
#>
#> Call:
#> roc.default(response = test.data$diabetes, predictor = predictions, ci = TRUE)
#>
#> Data: predictions in 145 controls (test.data$diabetes neg) < 73 cases (test.data$diabetes pos).
#> Area under the curve: 0.9345
#> 95% CI: 0.8969-0.9721 (DeLong)
Now I would like to consider out of fold predictions using the mlr3 package family according to this very helpful post: Tuning a stacked learner
#library
library(mlr3)
library(mlr3learners)
library(mlr3pipelines)
library(mlr3filters)
library(mlr3tuning)
library(paradox)
library(glmnet)
# creat elastic net regression
glmnet_lrn = lrn("classif.cv_glmnet", predict_type = "prob")
# create the learner out-of-bag predictions
glmnet_cv1 = po("learner_cv", glmnet_lrn, id = "glmnet") #I could not find a setting to filter the predictors (ie, not send the superdoc predictor here)
# summarize steps
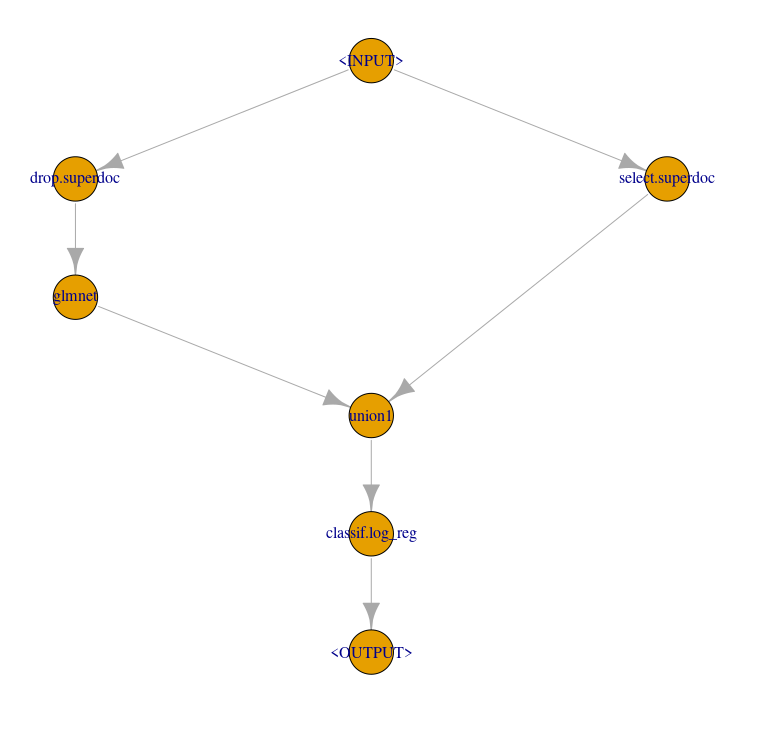
level0 = gunion(list(
glmnet_cv1,
po("nop", id = "only_superdoc_predictor"))) %>>% #I could not find a setting to send only the superdoc predictor to "union1"
po("featureunion", id = "union1")
# final logistic regression
log_reg_lrn = lrn("classif.log_reg", predict_type = "prob")
# combine ensemble model
ensemble = level0 %>>% log_reg_lrn
ensemble$plot(html = FALSE)

Created on 2021-03-15 by the reprex package (v1.0.0)
My question (I am rather new to the mlr3 package family)
- is the
mlr3package family well suited for the ensemble model I try to build? - if yes, how cold I finalize the ensemle model and make predictions on the
test.data

mlr3package familiy is very helpful. I read thatclassif.cv_glmnetdoes an internal 10-fold CV optimization. Can this somehow be repeated (repeatedcv) within the solution you proposed (as in mycaret::trainControlapproach) to avoid variability in smaller data sets? I think tuning both, alpha and lambda usingmlr3is not (yet) possible? – Grocer