thanks for your help. I'm trying to write an R function that will take a list containing numeric vectors and merge all the list elements which share numbers. I'm not sure I'm explaining the problem properly, so I hope you don't mind if I use an analogy. An example list could look like this:
> list(c(1, 6), c(2, 3), c(3, 2), c(4, 5, 6), c(5, 4), c(1, 6, 4))
[[1]]
[1] 1 6
[[2]]
[1] 2 3
[[3]]
[1] 3 2
[[4]]
[1] 4 5 6
[[5]]
[1] 5 4
[[6]]
[1] 1 6 4
If you imagine 6 villages, the list would show which villages are connected by roads. So list element [[1]] shows that village 1 is connected to village 1 and village 6. List element [[6]] shows that 6 is connected to village 1, village 6, and village 4. And so on. I want my output to show which villages are connected by the same 'road network', so village 1 is clearly in the same network as 6, but it should also be grouped with 4 and 5, because it is connected to them through 6 and then 4. 2 and 3 should then be grouped seperately, as they do not share a connection to the other network.
I've managed to piece together a solution, but it is deeply inelegant, and takes far too long to run for more complicated inputs. My solution is this:
input <- list(c(1, 6), c(2, 3), c(3, 2), c(4, 5, 6), c(5, 4), c(1, 6, 4))
remaining <- 1:6 # counter where i can store which numbers have not yet been evaluated
output <- vector("list", 6)
branch <- function(x) { # function to recursively evaluate vector elements
for(y in x) { # repeat for each vector element
if(y %in% remaining) { # check if the list element corresponding to y has been evaluated
output[[i]] <- append(output[[i]], input[[y]]) # assign list element y to output element i
assign("output", output, envir = globalenv()) #assign output to global environment
remaining <- remaining[remaining != y] # remove y from future evaluations
assign("remaining", remaining, envir = globalenv()) # assign remaining to global environment
branch(input[[y]]) # evaluate branches further from y
}
}
}
for(i in 1:6) { # repeat for each element of list
if(i %in% remaining) { # check if list element i has already been evaluated
branch(input[[i]]) # evaluate list element
}
}
output <- output[-which(sapply(output, is.null))] # remove null elements from list
output <- lapply(output, unique) # remove redundant elements from vectors
output
> output
[[1]]
[1] 1 6 4 5
[[2]]
[1] 2 3
Sorry for the long question, but I feel like there must be a simpler way to do this that I'm missing. Is anyone able to help out?

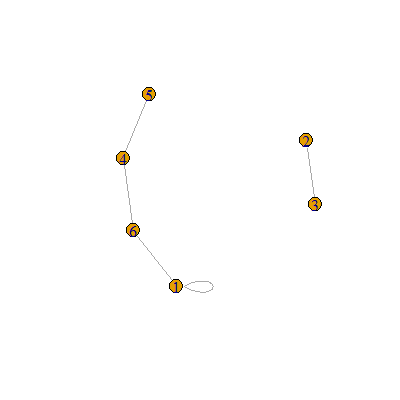
igraphwill make your analysis much easier, you just need to move your data intoigraph's required format first. – Horrocks