Out of this SO post resulted a discussion when benchmarking the various solutions. Consider the following code
# global environment is empty - new session just started
# set up
set.seed(20181231)
n <- sample(10^3:10^4,10^3)
for_loop <- function(n) {
out <- integer(length(n))
for(k in 1:length(out)) {
if((k %% 2) == 0){
out[k] <- 0L
next
}
out[k] <- 1L
next
}
out
}
# benchmarking
res <- microbenchmark::microbenchmark(
for_loop = {
out <- integer(length(n))
for(k in 1:length(out)) {
if((k %% 2) == 0){
out[k] <- 0L
next
}
out[k] <- 1L
next
}
out
},
for_loop(n),
times = 10^4
)
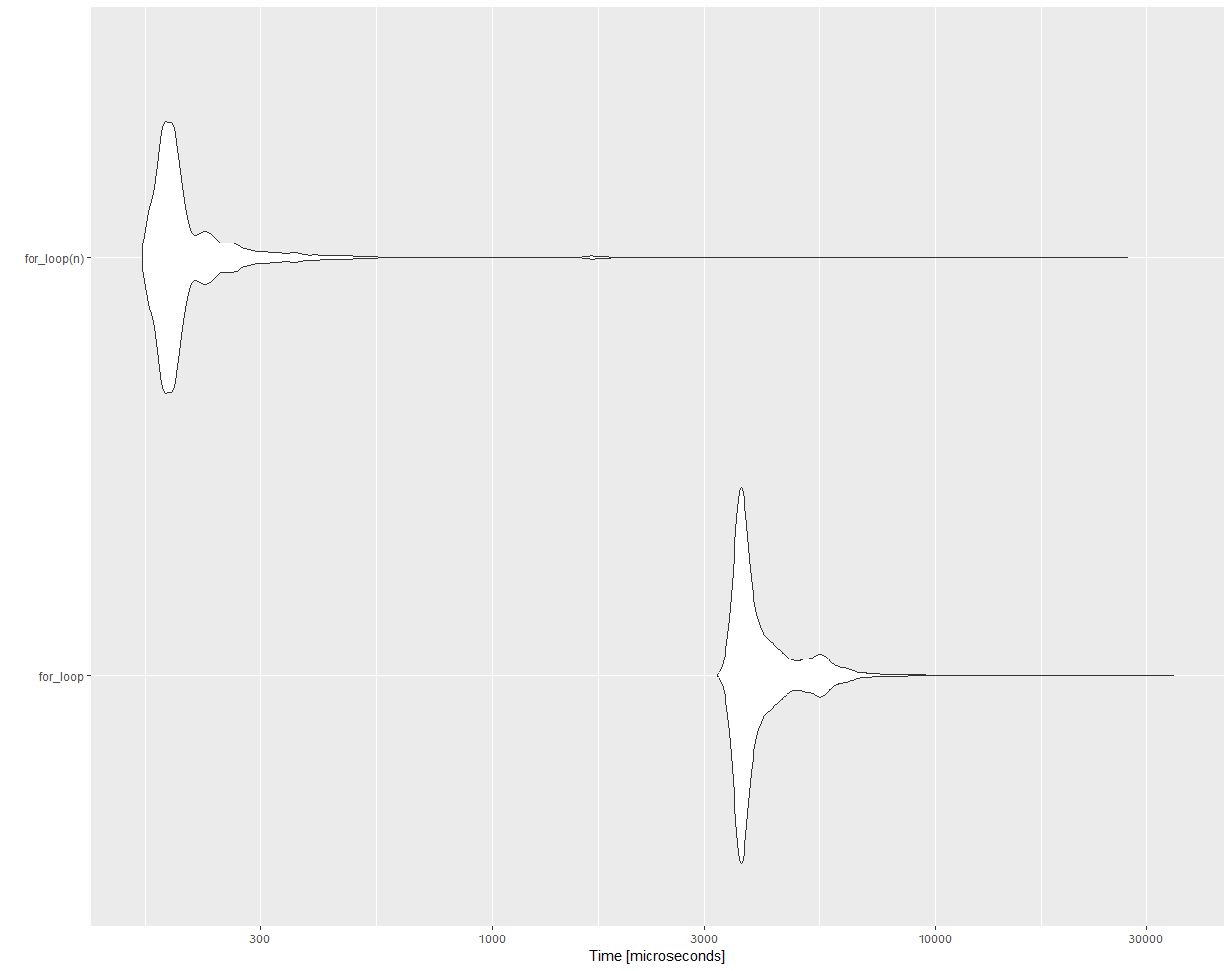
Here are the benchmarking results for the exact same loops, one packed in a function, the other not
# Unit: microseconds
# expr min lq mean median uq max neval cld
# for_loop 3216.773 3615.360 4120.3772 3759.771 4261.377 34388.95 10000 b
# for_loop(n) 162.280 180.149 225.8061 190.724 211.875 26991.58 10000 a
ggplot2::autoplot(res)
As can be seen, there is a drastic difference in efficiency. What is the underlying reason for this?
To be clear, the question is not about the task solved by the above code (which could be done much more elegantly) but merely about efficiency discrepancy between a regular loop and a loop wrapped inside a function.

for_loop. Do you believe it could still be the reason behind that lag? – Holusbolus