Per default, for the lower, middle and upper quantile in geom_boxplot the 25%-, 50%-, and 75%-quantiles are considered. These are computed from y, but can be set manually via the aesthetic arguments lower, upper, middle (providing also x, ymin and ymax and setting stat="identity").
However, doing so, several undesirable effects occur (cf. version 1 in the example code):
- The argument
groupis ignored, so all values of a column are considered in calculations (for instance when computing the lowest quantile for each group) - The resulting identical boxplots are grouped by
x, and repeated within the group as often as the specific group value occurs in the data (instead of merging the boxes to a wider one) - outliers are not plotted
By pre-computing the desired values and storing them in a new data frame, one can handle the first two points (cf. version 2 in the example code), while the third point is fixed by identifying the outliers and adding them separately to the chart via geom_point.
Is there a more straight forward way to have the quantiles changed, without having these undesired effects?
Example Code:
set.seed(12)
# Random data in B, grouped by values 1 to 4 in A
u <- data.frame(A = sample.int(4, 100, replace = TRUE), B = rnorm(100))
# Desired arguments
qymax <- 0.9
qymin <- 0.1
qmiddle <- 0.5
qupper <- 0.8
qlower <- 0.2
Version 1: Repeated boxplots per value in A, grouped by A
ggplot(u, aes(x = A, y = B)) +
geom_boxplot(aes(group=A,
lower = quantile(B, qlower),
upper = quantile(B, qupper),
middle = quantile(B, qmiddle),
ymin = quantile(B, qymin),
ymax = quantile(B, qymax) ),
stat="identity")
Version 2: Compute the arguments first for each group. Base R solution
Bgrouped <- lapply(unique(u$A), function(a) u$B[u$A == a])
.lower <- sapply(Bgrouped, function(x) quantile(x, qlower))
.upper <- sapply(Bgrouped, function(x) quantile(x, qupper))
.middle <- sapply(Bgrouped, function(x) quantile(x, qmiddle))
.ymin <- sapply(Bgrouped, function(x) quantile(x, qymin))
.ymax <- sapply(Bgrouped, function(x) quantile(x, qymax))
u <- data.frame(A = unique(u$A),
lower = .lower,
upper = .upper,
middle = .middle,
ymin = .ymin,
ymax = .ymax)
ggplot(u, aes(x = A)) +
geom_boxplot(aes(lower = lower, upper = upper,
middle = middle, ymin = ymin, ymax = ymax ),
stat="identity")

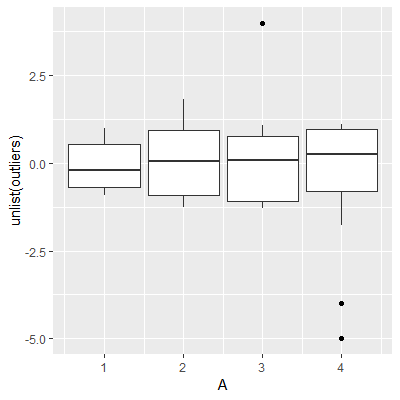
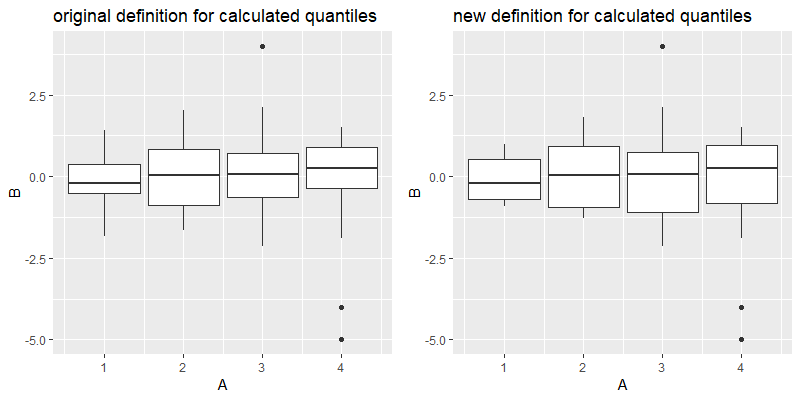
compute_grouphas to be modified next time I wish to change the default Parameters of ageomorstat. – Madigan