I'm trying to compute the 2 major principal components from a dataset in C++ with Eigen.
The way I do it at the moment is to normalize the data between [0, 1] and then center the mean. After that I compute the covariance matrix and run an eigenvalue decomposition on it. I know SVD is faster, but I'm confused about the computed components.
Here is the major code about how I do it (where traindata is my MxN sized input matrix):
Eigen::VectorXf normalize(Eigen::VectorXf vec) {
for (int i = 0; i < vec.size(); i++) { // normalize each feature.
vec[i] = (vec[i] - minCoeffs[i]) / scalingFactors[i];
}
return vec;
}
// Calculate normalization coefficients (globals of type Eigen::VectorXf).
maxCoeffs = traindata.colwise().maxCoeff();
minCoeffs = traindata.colwise().minCoeff();
scalingFactors = maxCoeffs - minCoeffs;
// For each datapoint.
for (int i = 0; i < traindata.rows(); i++) { // Normalize each datapoint.
traindata.row(i) = normalize(traindata.row(i));
}
// Mean centering data.
Eigen::VectorXf featureMeans = traindata.colwise().mean();
Eigen::MatrixXf centered = traindata.rowwise() - featureMeans;
// Compute the covariance matrix.
Eigen::MatrixXf cov = centered.adjoint() * centered;
cov = cov / (traindata.rows() - 1);
Eigen::SelfAdjointEigenSolver<Eigen::MatrixXf> eig(cov);
// Normalize eigenvalues to make them represent percentages.
Eigen::VectorXf normalizedEigenValues = eig.eigenvalues() / eig.eigenvalues().sum();
// Get the two major eigenvectors and omit the others.
Eigen::MatrixXf evecs = eig.eigenvectors();
Eigen::MatrixXf pcaTransform = evecs.rightCols(2);
// Map the dataset in the new two dimensional space.
traindata = traindata * pcaTransform;
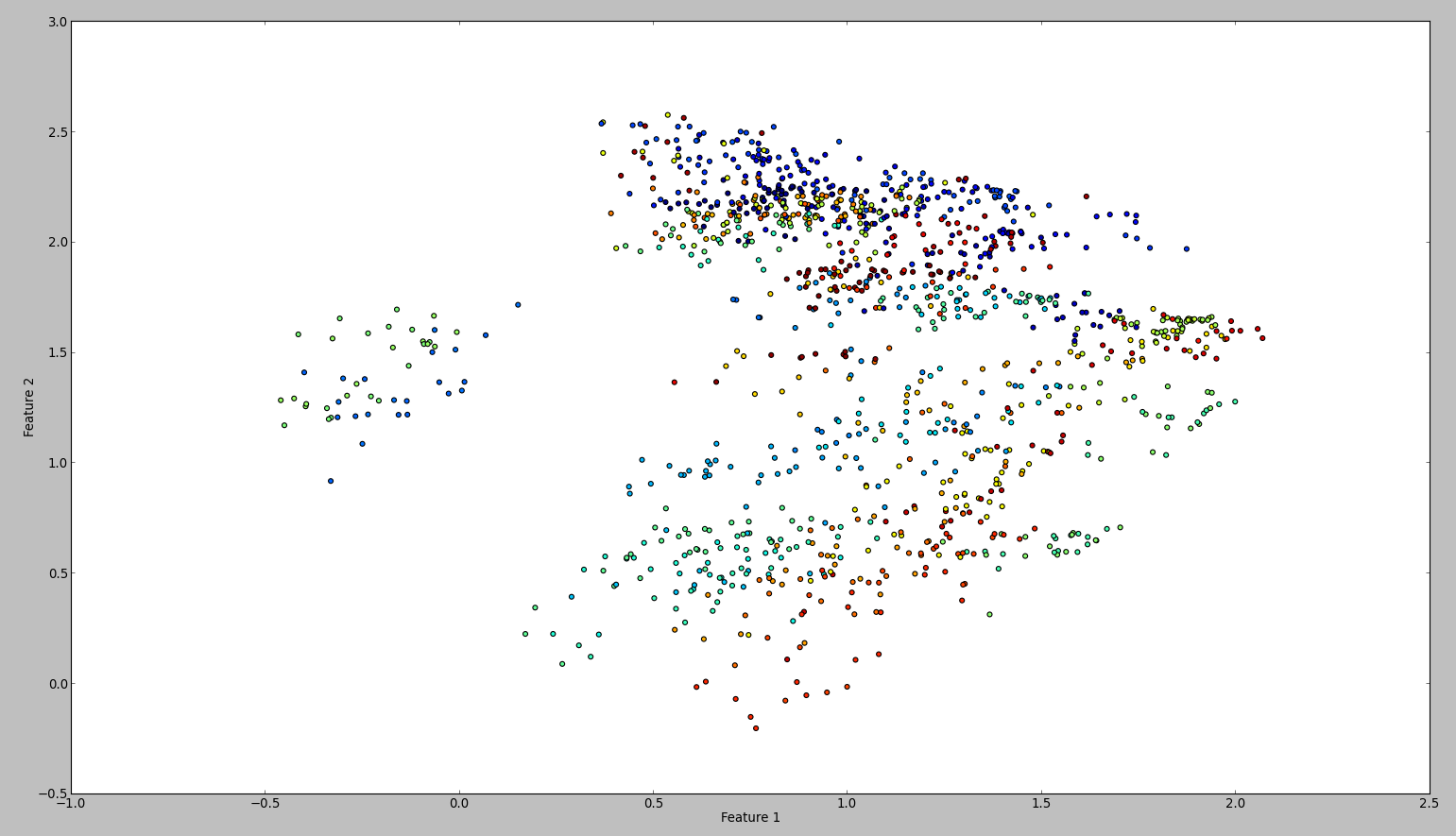
The result of this code is something like this:
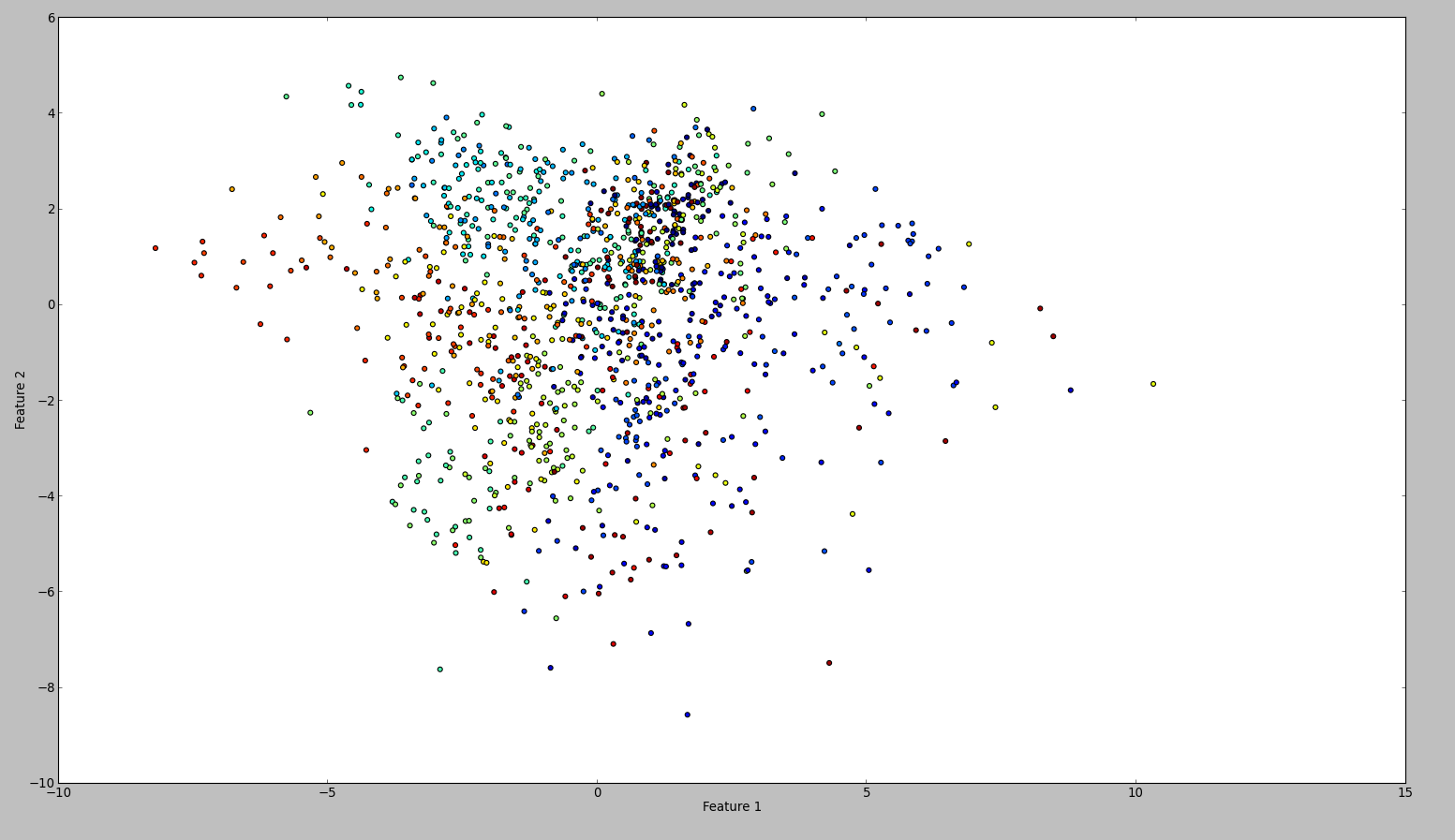
To confirm my results, I tried the same with WEKA. So what I did is to use the normalize and the center filter, in this order. Then the principal component filter and save + plot the output. The result is this:
Technically I should have done the same, however the outcome is so different. Can anyone see if I made a mistake?