dplyr >= 1.0.0
In newer versions of dplyr you can use rowwise() along with c_across to perform row-wise aggregation for functions that do not have specific row-wise variants, but if the row-wise variant exists it should be faster than using rowwise (eg rowSums, rowMeans).
Since rowwise() is just a special form of grouping and changes the way verbs work you'll likely want to pipe it to ungroup() after doing your row-wise operation.
To select a range by name:
df %>%
rowwise() %>%
mutate(sumrange = sum(c_across(x1:x5), na.rm = T))
# %>% ungroup() # you'll likely want to ungroup after using rowwise()
To select by type:
df %>%
rowwise() %>%
mutate(sumnumeric = sum(c_across(where(is.numeric)), na.rm = T))
# %>% ungroup() # you'll likely want to ungroup after using rowwise()
To select by column name:
You can use any number of tidy selection helpers like starts_with, ends_with, contains, etc.
df %>%
rowwise() %>%
mutate(sum_startswithx = sum(c_across(starts_with("x")), na.rm = T))
# %>% ungroup() # you'll likely want to ungroup after using rowwise()
To select by column index:
df %>%
rowwise() %>%
mutate(sumindex = sum(c_across(c(1:4, 5)), na.rm = T))
# %>% ungroup() # you'll likely want to ungroup after using rowwise()
rowise() will work for any summary function. However, in your specific case a row-wise variant exists (rowSums) so you can do the following, which will be faster:
df %>%
mutate(sumrow = rowSums(pick(x1:x5), na.rm = T))
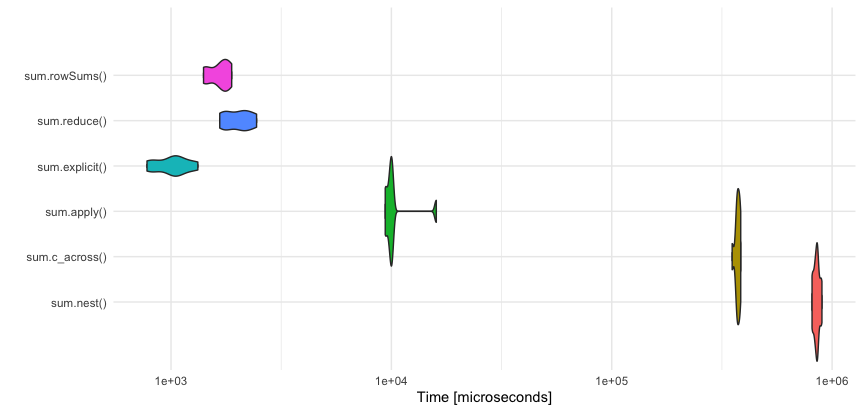
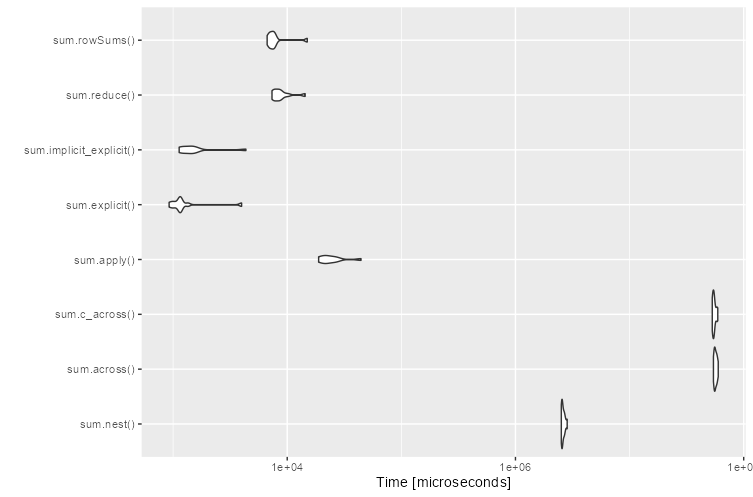
Benchmarking
rowwise makes a pipe chain very readable and works fine for smaller data frames. However, it is inefficient.
rowwise versus row-wise variant function
For this example, the the row-wise variant rowSums is much faster:
library(microbenchmark)
set.seed(1)
large_df <- slice_sample(df, n = 1E5, replace = T) # 100,000 obs
microbenchmark(
large_df %>%
rowwise() %>%
mutate(sumrange = sum(c_across(x1:x5), na.rm = T)),
large_df %>%
mutate(sumrow = rowSums(pick(x1:x5), na.rm = T)),
times = 10L
)
Unit: milliseconds
min lq mean median uq max neval cld
11108.459801 11464.276501 12144.871171 12295.362251 12690.913301 12918.106801 10 b
6.533301 6.649901 7.633951 7.808201 8.296101 8.693101 10 a
Large data frame without a row-wise variant function
If there isn't a row-wise variant for your function and you have a large data frame, consider a long-format, which is more efficient than rowwise. Though there are probably faster non-tidyverse options, here is a tidyverse option (using tidyr::pivot_longer):
library(tidyr)
tidyr_pivot <- function(){
large_df %>%
mutate(rn = row_number()) %>%
pivot_longer(cols = starts_with("x")) %>%
group_by(rn) %>%
summarize(std = sd(value, na.rm = T), .groups = "drop") %>%
bind_cols(large_df, .) %>%
select(-rn)
}
dplyr_rowwise <- function(){
large_df %>%
rowwise() %>%
mutate(std = sd(c_across(starts_with("x")), na.rm = T)) %>%
ungroup()
}
microbenchmark(dplyr_rowwise(),
tidyr_pivot(),
times = 10L)
Unit: seconds
expr min lq mean median uq max neval cld
dplyr_rowwise() 12.845572 13.48340 14.182836 14.30476 15.155155 15.409750 10 b
tidyr_pivot() 1.404393 1.56015 1.652546 1.62367 1.757428 1.981293 10 a
c_across versus pick
In the particular case of the sum function, pick and c_across give the same output for much of the code above:
sum_pick <- df %>%
rowwise() %>%
mutate(sumrange = sum(pick(x1:x5), na.rm = T))
sum_c_across <- df %>%
rowwise() %>%
mutate(sumrange = sum(c_across(x1:x5), na.rm = T))
all.equal(sum_pick, sum_c_across)
[1] TRUE
The row-wise output of c_across is a vector (hence the c_), while the row-wise output of pick is a 1-row tibble object:
df %>%
rowwise() %>%
mutate(c_across = list(c_across(x1:x5)),
pick = list(pick(x1:x5)),
.keep = "unused") %>%
ungroup()
# A tibble: 10 × 2
c_across pick
<list> <list>
1 <dbl [5]> <tibble [1 × 5]>
2 <dbl [5]> <tibble [1 × 5]>
3 <dbl [5]> <tibble [1 × 5]>
4 <dbl [5]> <tibble [1 × 5]>
5 <dbl [5]> <tibble [1 × 5]>
6 <dbl [5]> <tibble [1 × 5]>
7 <dbl [5]> <tibble [1 × 5]>
8 <dbl [5]> <tibble [1 × 5]>
9 <dbl [5]> <tibble [1 × 5]>
10 <dbl [5]> <tibble [1 × 5]>
The function you want to apply will necessitate, which verb you use. As shown above with sum you can use them nearly interchangeably. However, mean and many other common functions expect a (numeric) vector as its first argument:
class(df[1,])
"data.frame"
sum(df[1,]) # works with data.frame
[1] 4
mean(df[1,]) # does not work with data.frame
[1] NA
Warning message:
In mean.default(df[1, ]) : argument is not numeric or logical: returning NA
class(unname(unlist(df[1,])))
"numeric"
sum(unname(unlist(df[1,]))) # works with numeric vector
[1] 4
mean(unname(unlist(df[1,]))) # works with numeric vector
[1] 0.8
Ignoring the row-wise variant that exists for mean (rowMean) then in this case c_across should be used:
df %>%
rowwise() %>%
mutate(avg = mean(c_across(x1:x5), na.rm = T)) %>%
ungroup()
# A tibble: 10 x 6
x1 x2 x3 x4 x5 avg
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 0 1 1 0.8
2 0 1 1 0 1 0.6
3 0 NA 0 NA NA 0
4 NA 1 1 1 1 1
5 0 1 1 0 1 0.6
6 1 0 0 0 1 0.4
7 1 NA NA NA NA 1
8 NA NA NA 0 1 0.5
9 0 0 0 0 0 0
10 1 1 1 1 1 1
# Does not work
df %>%
rowwise() %>%
mutate(avg = mean(pick(x1:x5), na.rm = T)) %>%
ungroup()
rowSums, rowMeans, etc. can take a numeric data frame as the first argument, which is why they work with pick.

dplyr? Why not just a simpledf$sumrow <- rowSums(df, na.rm = TRUE)from base R? Ordf$sumrow <- Reduce(`+`, df)if you want to replicate the exact thing you did withdplyr. – Lavalleydplyrtoo as indf %>% mutate(sumrow = Reduce(`+`, .))ordf %>% mutate(sumrow = rowSums(.))– LavalleyrowSumsfunction works well. However, trying the suggestions indplyr;df %>% mutate(sumrow = rowSums(.))anddf %>% mutate(sumrow = Reduce(+, .))generate errors. I had just thought of fitting the operation within a dplyr chain alongside other preceding and subsequent operations in the chain.@DavidArenburg – Laryngitisdplyrversion and it will work. – Lavalleydplyrlike requested plus requires the exact same step of removingNAs byna.rm = TRUE. His second comment isdplyrbut (since theNAtreatment is missing) it leads toNAsums. – Ahona.rm = TRUEwhile your answer with an extrareplacestep and then using my exact comment is somewhat much better? Pretty sneaky (at best) statement and smells like rep wh*ring to me. – Lavalleydplyr, your data is probably not "tidy" and it might be better to reshape or to just use base. – Kexsummarise_each(funs(sum)), a comment to my answer pointed out that this was the right answer, so I improved it. I don't remeber if I even read your comment. Either way, I don't think this discussion is worth our time :) – Aho