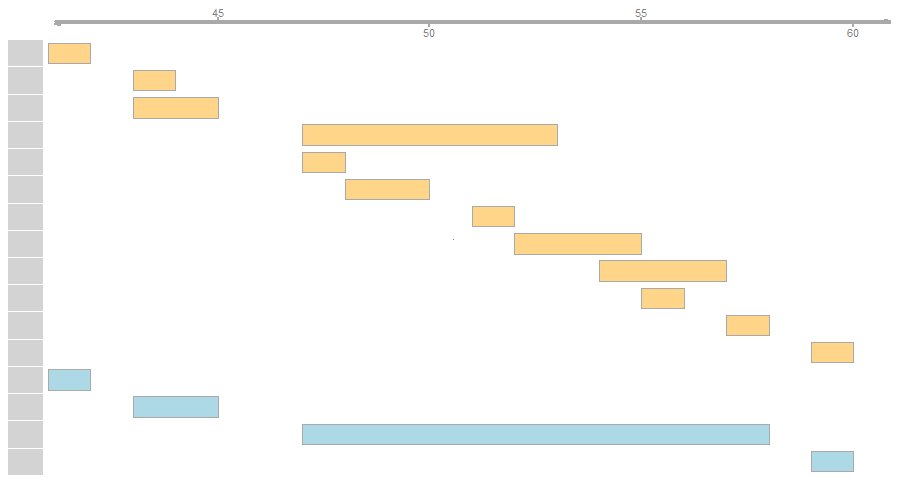
I need to aggregate overlapping segments into a single segment ranging all connected segments.
Note that a simple foverlaps cannot detect connections between non overlapping but connected segments, see the example for clarification. If it would rain on my segments in my plot I am looking for the stretches of dry ground.
So far I solve this problem by an iterative algorithm but I'm wondering if there is a more elegant and stright forward way for this problem. I'm sure not the first one to face it.
I was thinking about a non-equi rolling join, but faild to implement that
library(data.table)
(x <- data.table(start = c(41,43,43,47,47,48,51,52,54,55,57,59),
end = c(42,44,45,53,48,50,52,55,57,56,58,60)))
# start end
# 1: 41 42
# 2: 43 44
# 3: 43 45
# 4: 47 53
# 5: 47 48
# 6: 48 50
# 7: 51 52
# 8: 52 55
# 9: 54 57
# 10: 55 56
# 11: 57 58
# 12: 59 60
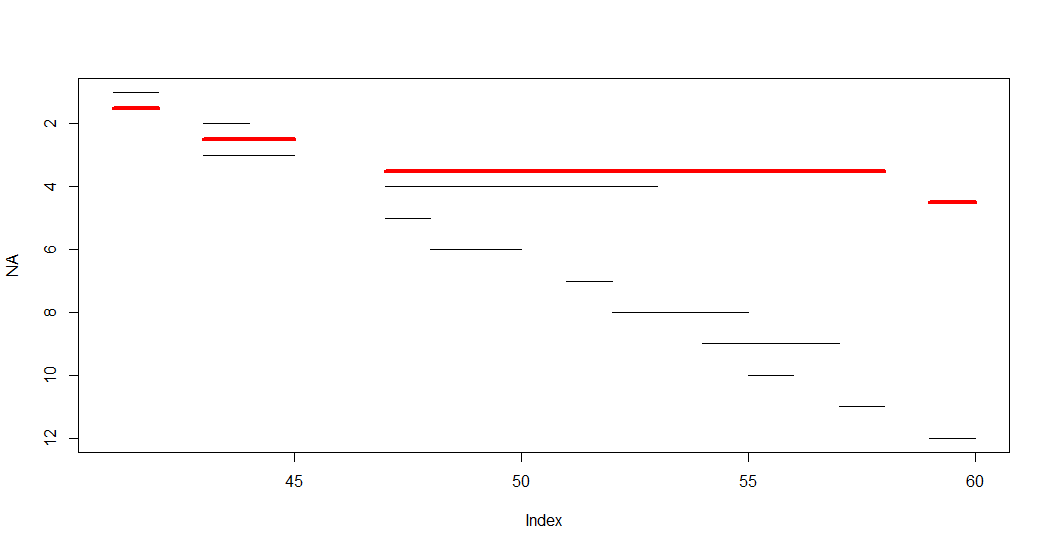
setorder(x, start)[, i := .I] # i is just a helper for plotting segments
plot(NA, xlim = range(x[,.(start,end)]), ylim = rev(range(x$i)))
do.call(segments, list(x$start, x$i, x$end, x$i))
x$grp <- c(1,3,3,2,2,2,2,2,2,2,2,4) # the grouping I am looking for
do.call(segments, list(x$start, x$i, x$end, x$i, col = x$grp))
(y <- x[, .(start = min(start), end = max(end)), k=grp])
# grp start end
# 1: 1 41 42
# 2: 2 47 58
# 3: 3 43 45
# 4: 4 59 60
do.call(segments, list(y$start, 12.2, y$end, 12.2, col = 1:4, lwd = 3))
EDIT:
That's brilliant, thanks, cummax & cumsum do the job, Uwe's Answer is slightly better than Davids comment.
end[.N]can get wrong results, try example dataxbelow.max(end)is correct in all cases, and faster.x <- data.table(start = c(11866, 12696, 13813, 14011, 14041), end = c(13140, 14045, 14051, 14039, 14045))min(start)andstart[1L]give the same (asxis ordered by start), the latter is faster.- grp on the fly is significantly faster, unfortunately I need grp assigned.
cumsum(cummax(shift(end, fill = 0)) < start)is significantly faster thancumsum(c(0, start[-1L] > cummax(head(end, -1L)))).- I did not test the package GenomicRanges solution.

x[, .(start[1L], end[.N]), by = .(grp = cumsum(c(0, start[-1L] > cummax(head(end, -1L)))))]could work. Basically a data.table version of my solution here – Fonzie