I have a 10 GB .dta Stata file and I am trying to read it into 64-bit R 3.3.1. I am working on a virtual machine with about 130 GB of RAM (4 TB HD) and the .dta file is about 3 million rows and somewhere between 400 and 800 variables.
I know data.table() is the fastest way to read in .txt and .csv files, but does anyone have a recommendation for reading largeish .dta files into R? Reading the file into Stata as a .dta file requires about 20-30 seconds, although I need to set my working memory max prior to opening the file (I set the max at 100 GB).
I have not tried importing to .csv in Stata, but I hope to avoid touching the file with Stata. A solution is found via Using memisc to import stata .dta file into R but this assumes RAM is scarce. In my case, I should have sufficient RAM to work with the file.

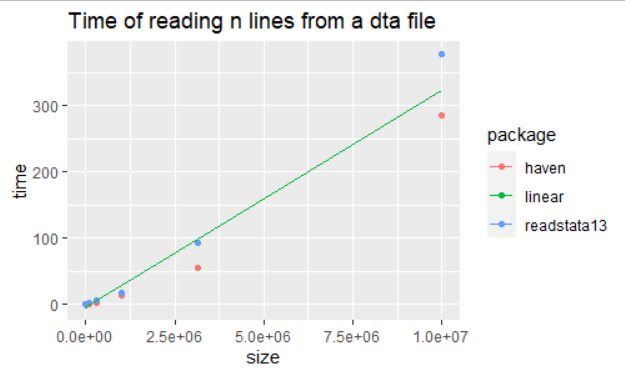
foreign::read.dta()should work, but it doesn't work on the latest stata format. – Izdta->csv->data.tablewould be your fastest option (although I hope not). If I were you I'd look through the results oflibrary(sos); findFn("stata dta")and benchmark on a reasonable (1GB?) size subset. – Gwendagwendolen