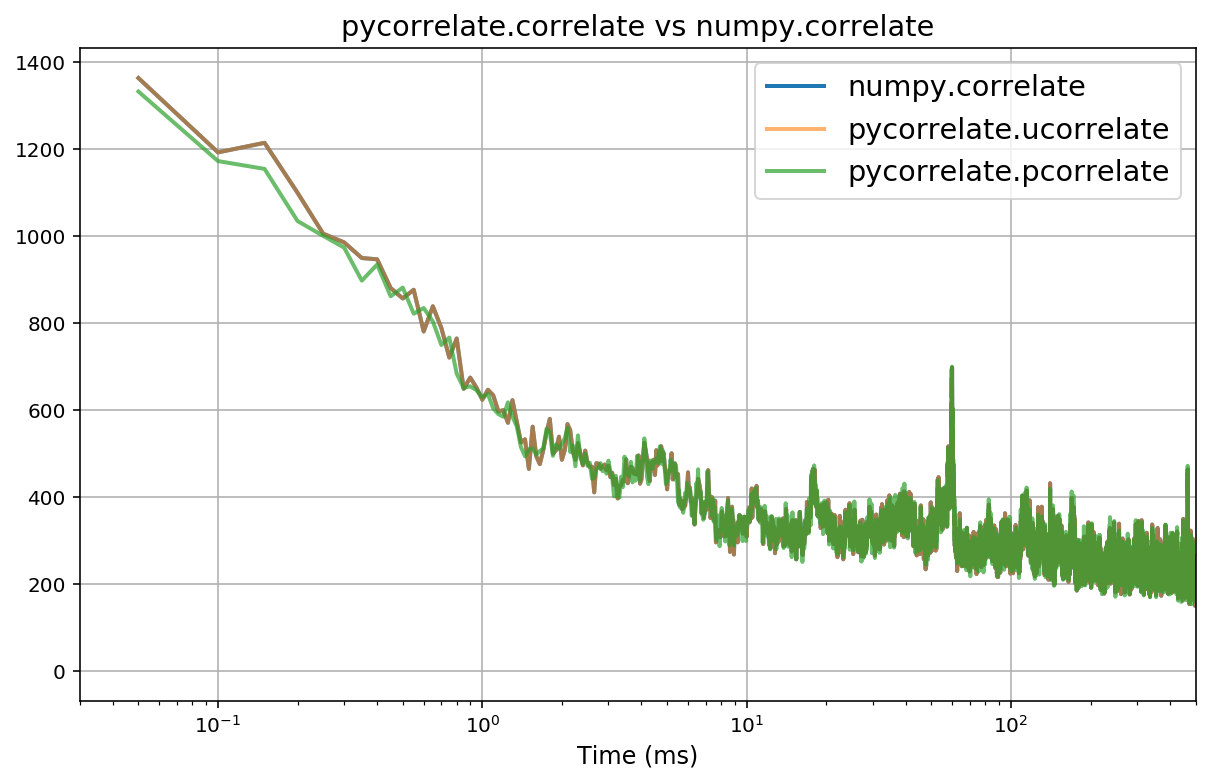
I am learning numpy/scipy, coming from a MATLAB background. The xcorr function in Matlab has an optional argument "maxlag" that limits the lag range from –maxlag to maxlag. This is very useful if you are looking at the cross-correlation between two very long time series but are only interested in the correlation within a certain time range. The performance increases are enormous considering that cross-correlation is incredibly expensive to compute.
In numpy/scipy it seems there are several options for computing cross-correlation. numpy.correlate, numpy.convolve, scipy.signal.fftconvolve. If someone wishes to explain the difference between these, I'd be happy to hear, but mainly what is troubling me is that none of them have a maxlag feature. This means that even if I only want to see correlations between two time series with lags between -100 and +100 ms, for example, it will still calculate the correlation for every lag between -20000 and +20000 ms (which is the length of the time series). This gives a 200x performance hit! Do I have to recode the cross-correlation function by hand to include this feature?