Suppose I have a numpy matrix like the following:
array([array([ 0.0072427 , 0.00669255, 0.00785213, 0.00845336, 0.01042869]),
array([ 0.00710799, 0.00668831, 0.00772334, 0.00777796, 0.01049965]),
array([ 0.00741872, 0.00650899, 0.00772273, 0.00729002, 0.00919407]),
array([ 0.00717589, 0.00627021, 0.0069514 , 0.0079332 , 0.01069545]),
array([ 0.00617369, 0.00590539, 0.00738468, 0.00761699, 0.00886915])], dtype=object)
How can I generate a 5 x 5 matrix where each index of the matrix is the cosine similarity of two corresponding rows in my original matrix?
e.g. row 0 column 2's value would be the cosine similarity between row 1 and row 3 in the original matrix.
Here's what I've tried:
from sklearn.metrics import pairwise_distances
from scipy.spatial.distance import cosine
import numpy as np
#features is a column in my artist_meta data frame
#where each value is a numpy array of 5 floating point values, similar to the
#form of the matrix referenced above but larger in volume
items_mat = np.array(artist_meta['features'].values)
dist_out = 1-pairwise_distances(items_mat, metric="cosine")
The above code gives me the following error:
ValueError: setting an array element with a sequence.
Not sure why I'm getting this because each array is of the same length (5), which I've verified.

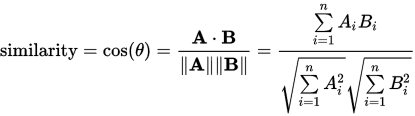
1-pairwise_distances(f,metric="cosine")is doing just what you need, assuming thatfis the original array at the top of your post. There may be an issue with the content ofitems_mat. Can you show the first, say, 5x5 elements of it? – Lewallenfis your matrix,1-pairwise_distances(f,metric="cosine")gives no errors whatsoever. – Lewallenfloatand notobject– Hemminger