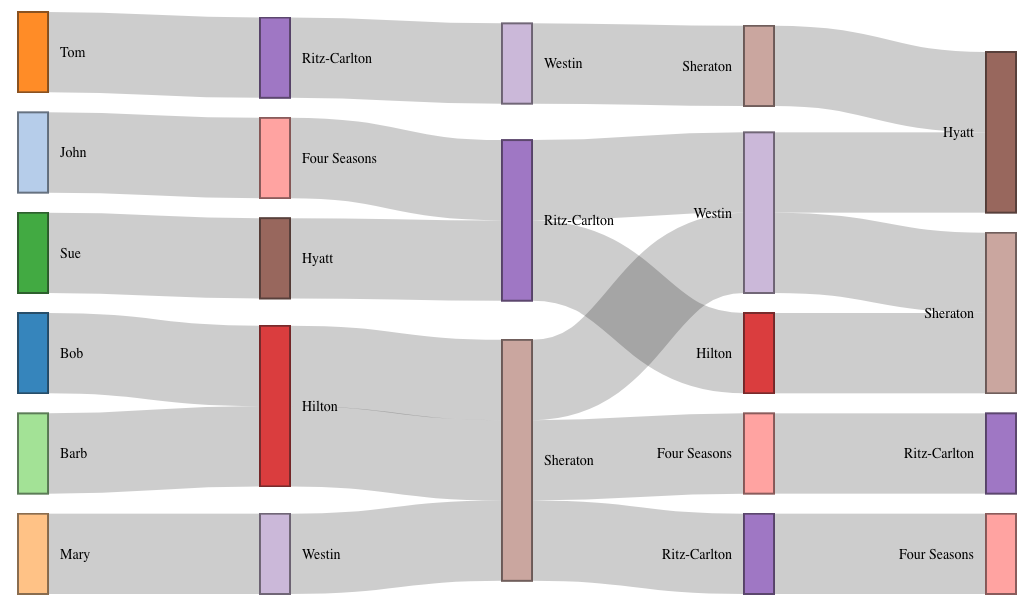
This question comes up a lot... how to convert a dataset that has multiple links/edges defined on each row across several columns. Here's how I convert that into the type of dataset that sankeyNetwork (and many other packages that deal with edges/links/network data) uses... a dataset with one edge/link per row.
starting with an example dataset...
df <- read.table(header = TRUE, stringsAsFactors = FALSE, text = '
name year1 year2 year3 year4
Bob Hilton Sheraton Westin Hyatt
John "Four Seasons" Ritz-Carlton Westin Sheraton
Tom Ritz-Carlton Westin Sheraton Hyatt
Mary Westin Sheraton "Four Seasons" Ritz-Carlton
Sue Hyatt Ritz-Carlton Hilton Sheraton
Barb Hilton Sheraton Ritz-Carlton "Four Seasons"
')
# name year1 year2 year3 year4
# 1 Bob Hilton Sheraton Westin Hyatt
# 2 John Four Seasons Ritz-Carlton Westin Sheraton
# 3 Tom Ritz-Carlton Westin Sheraton Hyatt
# 4 Mary Westin Sheraton Four Seasons Ritz-Carlton
# 5 Sue Hyatt Ritz-Carlton Hilton Sheraton
# 6 Barb Hilton Sheraton Ritz-Carlton Four Seasons
- create a row number so that you'll still be able to determine which row/observation each individual link came from when you convert the data to long format
- use
tidyr's pivot_longer() function to convert the dataset to long format
- convert the column name variable to the index/number of the column in the original dataset
- grouped by row (each observation in the original dataset), create a variable for each source node's "target" by setting it to the node following it in the next column
- filter out any rows that have
NA for "target" (nodes in the last column of the original dataset will not have a "target", and therefore those rows do not specify a link)
library(dplyr)
library(tidyr)
links <-
df %>%
mutate(row = row_number()) %>% # add a row id
pivot_longer(-row, names_to = "column", values_to = "source") %>% # gather all columns
mutate(column = match(column, names(df))) %>% # convert col names to col ids
group_by(row) %>%
mutate(target = lead(source, order_by = column)) %>% # get target from following node in row
ungroup() %>%
filter(!is.na(target)) # remove links from last column in original data
# # A tibble: 24 x 4
# row column source target
# <int> <int> <chr> <chr>
# 1 1 1 Bob Hilton
# 2 1 2 Hilton Sheraton
# 3 1 3 Sheraton Westin
# 4 1 4 Westin Hyatt
# 5 2 1 John Four Seasons
# 6 2 2 Four Seasons Ritz-Carlton
# 7 2 3 Ritz-Carlton Westin
# 8 2 4 Westin Sheraton
# 9 3 1 Tom Ritz-Carlton
# 10 3 2 Ritz-Carlton Westin
# # … with 14 more rows
Now the data is already in the typical network data format of one link per row defined by "source" and "target" columns, and it could be used with the sankeyNetwork(). However, you will likely want nodes referring to the same thing appearing multiple times within your plot... if someone visited the Hilton in year 1, and then visited the Hilton again in year 3, you will probably want 2 separate nodes, both named Hilton, but appearing in different parts of the plot. In order to do that, you will have to identify each node in your "source" and "target" columns with the year in which they were visited. That's where keeping the "row" and "column" variables around will come in handy.
Append the column index to the "source" name, and append the column index + 1 to the "target" name, and now you will be able to distinguish, for instance, between the node for Hilton which was visited in year 1 and the node for Hilton that was visited in year 3.
links <-
links %>%
mutate(source = paste0(source, '_', column)) %>%
mutate(target = paste0(target, '_', column + 1)) %>%
select(source, target)
# # A tibble: 24 x 2
# source target
# <chr> <chr>
# 1 Bob_1 Hilton_2
# 2 Hilton_2 Sheraton_3
# 3 Sheraton_3 Westin_4
# 4 Westin_4 Hyatt_5
# 5 John_1 Four Seasons_2
# 6 Four Seasons_2 Ritz-Carlton_3
# 7 Ritz-Carlton_3 Westin_4
# 8 Westin_4 Sheraton_5
# 9 Tom_1 Ritz-Carlton_2
# 10 Ritz-Carlton_2 Westin_3
# # … with 14 more rows
Now you can follow the rather standard procedure for using a source-target list of links to build the necessary data frames for sankeyNetwork().
Create a nodes data frame with all the unique nodes found in the "source" and "target" vectors. You can also create a label vector in the nodes data frame that does not include the year/column id suffix.
nodes <- data.frame(name = unique(c(links$source, links$target)))
nodes$label <- sub('_[0-9]*$', '', nodes$name) # remove column id from node label
# # A tibble: 23 x 2
# name label
# <chr> <chr>
# 1 Bob_1 Bob
# 2 Hilton_2 Hilton
# 3 Sheraton_3 Sheraton
# 4 Westin_4 Westin
# 5 John_1 John
# 6 Four Seasons_2 Four Seasons
# 7 Ritz-Carlton_3 Ritz-Carlton
# 8 Tom_1 Tom
# 9 Ritz-Carlton_2 Ritz-Carlton
# 10 Westin_3 Westin
# # … with 13 more rows
Convert the "source" and "target" vectors in the links data frame to be the 0-based-index of the node in the nodes data frame. Add an arbitrary value for each link in the links data frame since it's required by sankeyNetwork(). Then plot it with sankeyNetwork()!
links$source_id <- match(links$source, nodes$name) - 1
links$target_id <- match(links$target, nodes$name) - 1
links$value <- 1
library(networkD3)
sankeyNetwork(Links = links, Nodes = nodes, Source = 'source_id',
Target = 'target_id', Value = 'value', NodeID = 'label')
![enter image description here]()