I managed to create parallel connections in R to a TSQL server using the below code:
SQL_retrieve <- function(x){
con <- odbcDriverConnect(
'driver={SQL Server};server=OPTMSLMSOFT02;database=Ad_History;trusted_connection=true')
odbcGetInfo(con)
rawData <- sqlQuery(con,
paste("select * from AD_MDL_R_INPUT a where a.itm_lctn_num = ",
facility[x] )) odbcClose(con) return(rawData) }
cl <- makeCluster(5) registerDoParallel(cl)
outputPar <- foreach(j = 1:facility_count, .packages="RODBC")
%dopar% SQL_retrieve(j) stopCluster(cl)
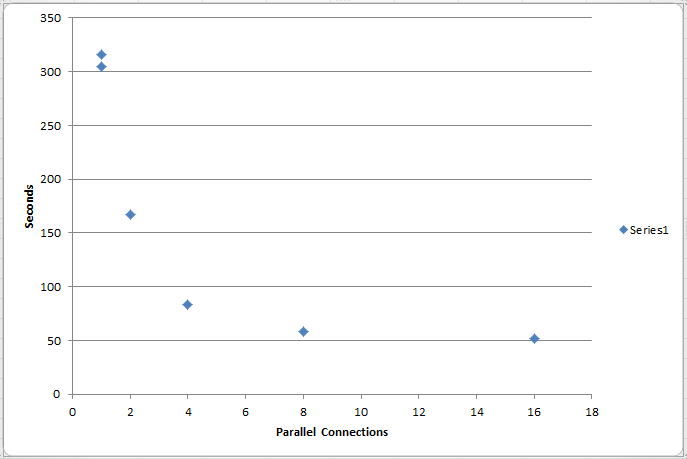
I would expect to see all connections actively downloading in parallel, but the reality is that only one or two connections are active at a time (see image below).
Even with 32 connections, the total download time is cut by slightly more than 1/2 (should be closer to 1/32, in theory, right?). There are also large pauses between connection activity. Why is this?
Some notes to keep in mind:
- The TSQL server and R are both on the same server, so network latency not an issue.
- The TSQL server allows up to a max of ~32k connections, so we are not bumping into a session limit issue.
UPDATE 7/26/17 Taking another stab at this problem and it now works (code unchanged). Not sure what happened between now and initial posting, but perhaps some changes to MS SQL server settings (unlikely).
The time to pull 7.9 million rows follows the curve in the image below.