I have 10 greyscale brain MRI scans from BrainWeb. They are stored as a 4d numpy array, brains, with shape (10, 181, 217, 181). Each of the 10 brains is made up of 181 slices along the z-plane (going through the top of the head to the neck) where each slice is 181 pixels by 217 pixels in the x (ear to ear) and y (eyes to back of head) planes respectively.
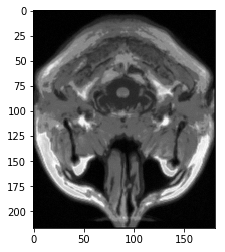
All of the brains are type dtype('float64'). The maximum pixel intensity across all brains is ~1328 and the minimum is ~0. For example, for the first brain, I calculate this by brains[0].max() giving 1328.338086605072 and brains[0].min() giving 0.0003886114541273855. Below is a plot of a slice of a brain[0]:
I want to binarize all these brain images by rescaling the pixel intensities from [0, 1328] to {0, 1}. Is my method correct?
I do this by first normalising the pixel intensities to [0, 1]:
normalized_brains = brains/1328
And then by using the binomial distribution to binarize each pixel:
binarized_brains = np.random.binomial(1, (normalized_brains))
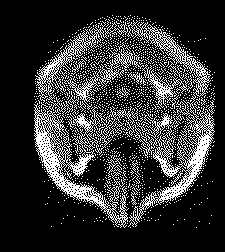
The plotted result looks correct:
A 0 pixel intensity represents black (background) and 1 pixel intensity represents white (brain).
I experimented by implementing another method to normalise an image from this post but it gave me just a black image. This is because np.finfo(np.float64) is 1.7976931348623157e+308, so the normalization step
normalized_brains = brains/1.7976931348623157e+308
just returned an array of zeros which in the binarizition step also led to an array of zeros.
Am I binarising my images using a correct method?