I need to reproduce plots generated in InDesign in ggplot for reproducibility.
In this particular example, I have two plots that are combined into one composite plot (I have used the package {patchwork} for this).
I then need to overlay lines joining key points on one plot with the corresponding points on the bottom plot.
The two plots are generated from the same data, have the same x-axis values, but different y-axis values.
I have seen these examples on Stack Overflow, but these deal with drawing lines across facets, which doesn't work here as I'm attempting to draw lines across separate plots:
I've tried several approaches, and my closest so far has been to:
- Add the lines with grobs using
{grid}package - Convert the second plot to a gtable using
{gtable}and set the clip of the panel to off so that I can extend the lines upwards beyond the panel of the plot. - Combine the plots again into a single image with
{patchwork}.
The problem comes in the last step as the x-axes now do not line up anymore as they did before adding the lines and setting the clip to off (see example in code).
I have also tried combining the plots with ggarrange, {cowplot} and {egg} and {patchwork} comes the closest.
Following is my attempt at the best minimal reprex I can create, but still capturing the nuances of what it is I want to achieve.
library(ggplot2)
library(dplyr)
library(tidyr)
library(patchwork)
library(gtable)
library(grid)
# DATA
x <- 1:20
data <- data.frame(
quantity = x,
curve1 = 10 + 50*exp(-0.2 * x),
curve2 = 5 + 50*exp(-0.5 * x),
profit = c(seq(10, 100, by = 10),
seq(120, -240, by = -40))
)
data_long <- data %>%
gather(key = "variable", value = "value", -quantity)
# POINTS AND LINES
POINTS <- data.frame(
label = c("B", "C"),
quantity = c(5, 10),
value = c(28.39397, 16.76676),
profit = c(50, 100)
)
GROB <- linesGrob()
# Set maximum y-value to extend lines to outside of plot area
GROB_MAX <- 200
# BASE PLOTS
# Plot 1
p1 <- data_long %>%
filter(variable != "profit") %>%
ggplot(aes(x = quantity, y = value)) +
geom_line(aes(color = variable)) +
labs(x = "") +
coord_cartesian(xlim = c(0, 20), ylim = c(0, 30), expand = FALSE) +
theme(legend.justification = "top")
p1
# Plot 2
p2 <- data_long %>%
filter(variable == "profit") %>%
ggplot(aes(x = quantity, y = value)) +
geom_line(color = "darkgreen") +
coord_cartesian(xlim = c(0, 20), ylim = c(-100, 120), expand = FALSE) +
theme(legend.position = "none")
p2
# PANEL A
panel_A <- p1 + p2 + plot_layout(ncol = 1)
panel_A
# PANEL B
# ATTEMPT - adding grobs to plot 1 that end at x-axis of p1
p1 <- p1 +
annotation_custom(GROB,
xmin = 0,
xmax = POINTS$quantity[POINTS$label == "B"],
ymin = POINTS$value[POINTS$label == "B"],
ymax = POINTS$value[POINTS$label == "B"]) +
annotation_custom(GROB,
xmin = POINTS$quantity[POINTS$label == "B"],
xmax = POINTS$quantity[POINTS$label == "B"],
ymin = 0,
ymax = POINTS$value[POINTS$label == "B"]) +
geom_point(data = POINTS %>% filter(label == "B"), size = 1)
# ATTEMPT - adding grobs to plot 2 that extend up to meet plot 1
p2 <- p2 + annotation_custom(GROB,
xmin = POINTS$quantity[POINTS$label == "B"],
xmax = POINTS$quantity[POINTS$label == "B"],
ymin = POINTS$profit[POINTS$label == "B"],
ymax = GROB_MAX)
# Create gtable from ggplot
g2 <- ggplotGrob(p2)
# Turn clip off for panel so that line can extend above
g2$layout$clip[g2$layout$name == "panel"] <- "off"
panel_B <- p1 + g2 + plot_layout(ncol = 1)
panel_B
# Problems:
# 1. Note the shift in axes when turning the clip off so now they do not line up anymore.
# 2. Turning the clip off mean plot 2 extends below the axis. Tried experimenting with various clips.
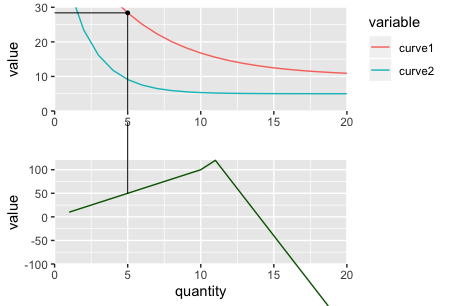
The expectation is that the plots in panel_B should still appear as they do in panel_A but have the joining lines linking points between the plots.
I am looking for help with solving the above, or else, alternative approaches to try out.
As a reference without running the code above - links to images as I can't post them.
Panel A
Panel B: What it currently looks like
Panel B: What I want it to look like!







ggplot. – Piggin