I want to compare some sub-groups of my data in one plot and some other sub-groups in another plot. If I make one plot with all sub-groups plotted, the figure is overwhelming and each individual comparison becomes difficult. I think it will make more sense to the reader if a given subgroup is the same color across all plots.
Here's are two things I've tried that almost work, but neither quite works. They're as close as I can come to a MWE!
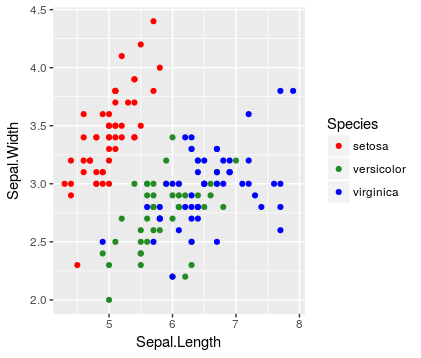
Wrong because all three levels are shown in the legend
library(tidyverse)
# compare first and second species
ggplot(data = iris %>% filter(Species != 'virginica'),
mapping = aes(x = Sepal.Length,
y = Sepal.Width,
color = Species)) +
geom_point() +
scale_color_discrete(drop = FALSE)
# compare second and third species
ggplot(data = iris %>% filter(Species != 'setosa'),
mapping = aes(x = Sepal.Length,
y = Sepal.Width,
color = Species)) +
geom_point() +
scale_color_discrete(drop = FALSE)


Note that the un-plotted level still appears in the legend (consistent with the idea of drop = FALSE).
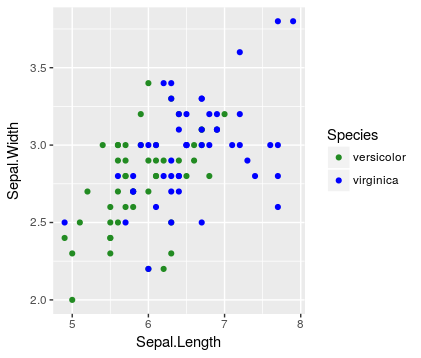
Wrong because the second plot doesn't maintain the species-color mapping established by the first plot
# compare first and second species
ggplot(data = iris %>% filter(Species != 'virginica'),
mapping = aes(x = Sepal.Length,
y = Sepal.Width,
color = Species)) +
geom_point() +
scale_color_manual(values = c('red', 'forestgreen', 'blue'),
breaks = unique(iris$Species))
# compare second and third species
ggplot(data = iris %>% filter(Species != 'setosa'),
mapping = aes(x = Sepal.Length,
y = Sepal.Width,
color = Species)) +
geom_point() +
scale_color_manual(values = c('red', 'forestgreen', 'blue'),
breaks = unique(iris$Species))


Note that in the left plot setosa = red and virginica = green, but in the right plot that mapping is changed.