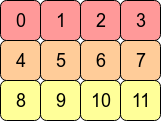
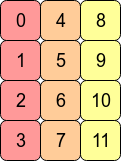
Maybe this example with 12 different array values will help:
In [207]: x=np.arange(12).reshape(3,4).copy()
In [208]: x.flags
Out[208]:
C_CONTIGUOUS : True
F_CONTIGUOUS : False
OWNDATA : True
...
In [209]: x.T.flags
Out[209]:
C_CONTIGUOUS : False
F_CONTIGUOUS : True
OWNDATA : False
...
The C order values are in the order that they were generated in. The transposed ones are not
In [212]: x.reshape(12,) # same as x.ravel()
Out[212]: array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11])
In [213]: x.T.reshape(12,)
Out[213]: array([ 0, 4, 8, 1, 5, 9, 2, 6, 10, 3, 7, 11])
You can get 1d views of both
In [214]: x1=x.T
In [217]: x.shape=(12,)
the shape of x can also be changed.
In [220]: x1.shape=(12,)
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
<ipython-input-220-cf2b1a308253> in <module>()
----> 1 x1.shape=(12,)
AttributeError: incompatible shape for a non-contiguous array
But the shape of the transpose cannot be changed. The data is still in the 0,1,2,3,4... order, which can't be accessed accessed as 0,4,8... in a 1d array.
But a copy of x1 can be changed:
In [227]: x2=x1.copy()
In [228]: x2.flags
Out[228]:
C_CONTIGUOUS : True
F_CONTIGUOUS : False
OWNDATA : True
...
In [229]: x2.shape=(12,)
Looking at strides might also help. A strides is how far (in bytes) it has to step to get to the next value. For a 2d array, there will be be 2 stride values:
In [233]: x=np.arange(12).reshape(3,4).copy()
In [234]: x.strides
Out[234]: (16, 4)
To get to the next row, step 16 bytes, next column only 4.
In [235]: x1.strides
Out[235]: (4, 16)
Transpose just switches the order of the strides. The next row is only 4 bytes- i.e. the next number.
In [236]: x.shape=(12,)
In [237]: x.strides
Out[237]: (4,)
Changing the shape also changes the strides - just step through the buffer 4 bytes at a time.
In [238]: x2=x1.copy()
In [239]: x2.strides
Out[239]: (12, 4)
Even though x2 looks just like x1, it has its own data buffer, with the values in a different order. The next column is now 4 bytes over, while the next row is 12 (3*4).
In [240]: x2.shape=(12,)
In [241]: x2.strides
Out[241]: (4,)
And as with x, changing the shape to 1d reduces the strides to (4,).
For x1, with data in the 0,1,2,... order, there isn't a 1d stride that would give 0,4,8....
__array_interface__ is another useful way of displaying array information:
In [242]: x1.__array_interface__
Out[242]:
{'strides': (4, 16),
'typestr': '<i4',
'shape': (4, 3),
'version': 3,
'data': (163336056, False),
'descr': [('', '<i4')]}
The x1 data buffer address will be same as for x, with which it shares the data. x2 has a different buffer address.
You could also experiment with adding a order='F' parameter to the copy and reshape commands.