Ok, here is yet another missing value filling question.
I am looking for a way to fill NAs based on both the previous and next existent values in a column. Standard filling in a single direction is not sufficient for this task.
If the previous and next valid values in a column are not the same, then the chunk remains as NA.
The code for the sample data frame is:
df_in <- tibble(id= 1:12,
var1 = letters[1:12],
var2 = c(NA,rep("A",2),rep(NA,2),rep("A",2),rep(NA,2),rep("B",2),NA))
Thanks,

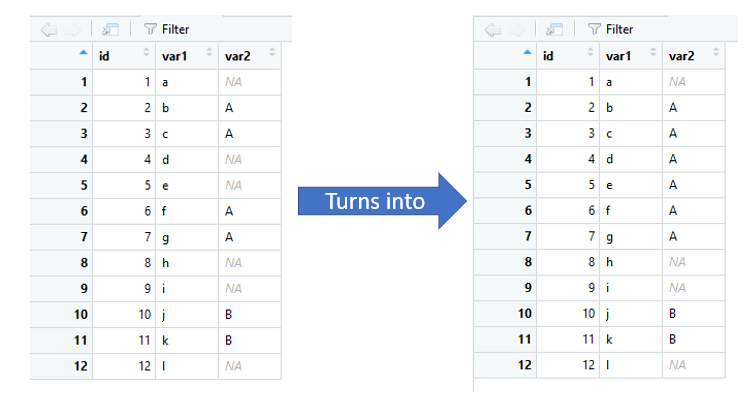
Aand the next valueNA. Therefore it should stayNA. Similarly for row 5. Could you please clarify? – KefferNA' here. – Mopboard