Numpy 1.25
Correcting the initial approach
First, let's make the initial approach work. There are two important parts: splitting the data into diagonals and merging them back together.
The smallest and largest diagonal offsets are -(hight-1) and width-1 respectively. So we can simplify splitting into diagonals as follows:
def get_diagonals(array):
'''Return all the top-right to bottom-left diagonals of an array
starting from the top-left corner.
'''
height, width = array.shape
fliplr_arr = np.fliplr(array)
return [fliplr_arr.diagonal(offset).copy() # Starting in NumPy 1.9 diagonal returns a read-only view on the original array
for offset in range(width-1, -height, -1)]
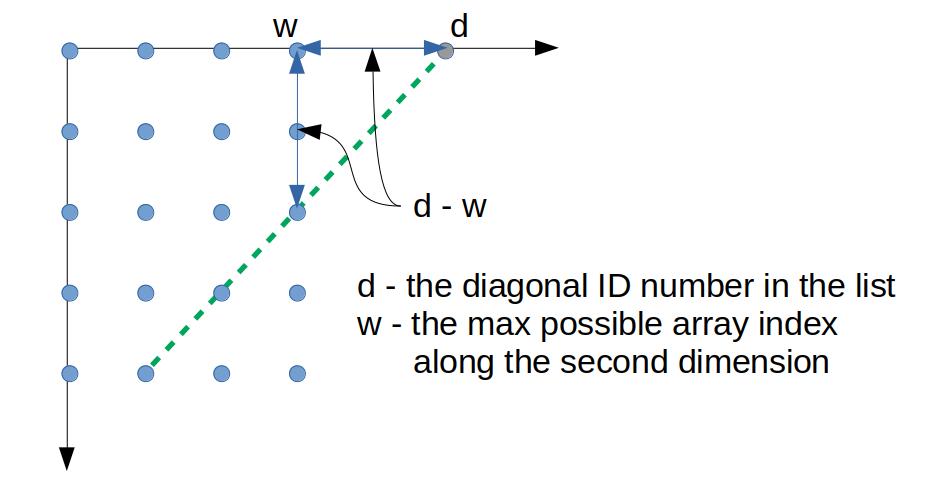
Having all diagonals collected in a list, let's numerate them by their index. Now, for all diagonals with index less then the array width, the index of an element within the diagonal is equal to its first index within the array. For other diagonals, a position of an element within the diagonal plus the height of the diagonal starting point gives the height of the element within the array. The diagonal starting point is equal to the difference of the diagonal number and the last possible index within the second array dimension (see the picture below).
![figure 1]()
So we can rewrite the second part of the code as follows:
def from_diagonals(diagonals, shape, dtype):
arr = np.empty(shape, dtype)
height, width = shape
for h in range(height):
arr[h, :] = [
diagonal[h if d < width else h - (d - (width-1))]
for d, diagonal in enumerate(diagonals[h:h+width], start=h)
]
return arr
Now the whole process of work is as follows:
arr = ...
diagonals = get_diagonals(arr)
for diag in diagonals:
...
new_arr = from_diagonals(diagonals, arr.shape, arr.dtype)
Working with indexes
We can create a function to reach data by the diagonal indexes like this:
def get_diagonal_function(height, width, base=0, antidiagonal=False):
from functools import partial
index = np.mgrid[:height, :width]
if antidiagonal:
index = index[..., ::-1] # flip horizontal indices left-to-right
_diag = partial(index.diagonal, axis1=1, axis2=2)
max_offset = width - 1
shift = max_offset + base
diagonal = lambda x: _diag(shift - x)
diagonal.min = base
diagonal.max = (height-1) + (width-1) + base
diagonal.off = _diag
return diagonal
Example of extracting all anti-diagonals:
diagonal = get_diagonal_function(*arr.shape, base=1, antidiagonal=True)
diagonals = [arr[*diagonal(x)] for x in range(diagonal.min, 1+diagonal.max)]
main_antidiagonal = diagonal.off(0)
Example of flipping data along anti-diagonals:
arr = [
[ 1, 2, 4],
[ 3, 5, 7],
[ 6, 8, 10],
[ 9, 11, 13],
[12, 14, 15]
]
arr = np.array(arr)
diagonal = get_diagonal_function(*arr.shape, antidiagonal=True)
for x in range(1+diagonal.max):
index = tuple(diagonal(x))
arr[index] = arr[index][::-1]
print(arr)
[[ 1 3 6]
[ 2 5 9]
[ 4 8 12]
[ 7 11 14]
[10 13 15]]
Converting coordinates
Warning: The code below has not been properly tested; it is not intended for fast calculations; it is just an ad hoc proof of concept.
In case of anti-diagonals as described in the original post, converting can be performed as follows (see figure above for reference):
def diag_to_array(d, x, shape, direction='top-bottom,right-left'):
height, width = shape
h = x if d < width else x + (d - (width-1))
w = d - h
return h, w
In general, converting resembles an affine transformation (with cutting at the edges, if I may say so). But we need to know the direction and ordering of diagonals, of which there can be 16 options. They can be given by vectors indicating the direction of counting diagonals and elements along them. These vectors can also be used to determine a new origin.
def get_origin(d, e, shape):
height, width = np.array(shape) - 1 # max coordinate values along dimensions
match d, e:
case ((1, x), (y, 1)) | ((x, 1), (1, y)):
origin = (0, 0)
case ((1, x), (y, -1)) | ((x, -1), (1, y)):
origin = (0, width)
case ((-1, x), (y, 1)) | ((x, 1), (-1, y)):
origin = (height, 0)
case ((-1, x), (y, -1)) | ((x, -1), (-1, y)):
origin = (height, width)
return np.array(origin)
Notes:
d is one of the {(1, 0), (-1, 0), (0, 1), (0, -1)} vectors showing the direction of the diagonal number coordinate;e is one of the {(1, 1), (1, -1), (-1, 1), (-1, -1)} vectors showing the direction of numbering elements along the diagonal.
As for coordinate transformations, it seems more convenient to implement them in a class:
class Transfomer:
def __init__(self, d, e, shape):
self.d = np.array(d)
self.e = np.array(e)
self.shape = np.array(shape)
self.A = np.stack([self.d, self.e])
self.origin = get_origin(d, e, shape)
self.edge = abs(self.d @ self.shape) - 1
def diag_to_array(self, ndiagonal, element):
'''Return the array coordiantes where:
ndiagonal: the number of a diagonal
element: the element index on the diagonal
'''
if ndiagonal > self.edge:
element = element + ndiagonal - self.edge
elif ndiagonal < 0:
element = element - ndiagonal
diag_coords = np.array([ndiagonal, element])
array_coords = diag_coords @ self.A + self.origin
return array_coords
def array_to_diag(self, *args, **kwargs):
raise NotImplementedError
Example:
d = (0, 1) # take anti-diagonals from left to right starting from the top-left corner
e = (1, -1) # and index their elements from top-right to bottom-left
arr = np.arange(5*3).reshape(5, 3)
coords = Transfomer(d, e, arr.shape)
diagonal = get_diagonal_function(*arr.shape, base=1, antidiagonal=True)
diagonals = [arr[*diagonal(x)] for x in range(diagonal.min, 1+diagonal.max)]
for ndiag, element in [(0, 0), (2, 1), (3, 0), (5, 1), (6, 0)]:
array_point = coords.diag_to_array(ndiag, element)
try:
assert diagonals[ndiag][element] == arr[*array_point], f'{ndiag=}, {element=}'
except AssertionError as e:
print(e)
Working with diagonals as a writeable view
If the array structure is straight simple (i.e. a contiguous sequence of data with no stride tricks or complicated transpositions), we can try (with some caution) to force a diagonal view to be writeable. Then all changes will be made with the original data, and you don't have to reassemble the diagonals again. Let's do mirroring anti-diagonals, for example:
def diag_range(shape, asnumpy=False):
'''Returns a sequence of numbers arranged diagonally
in a given shape as separate diagonals
'''
diagonals = []
dmin, dmax = min(shape), max(shape)
start = 1
for n in range(dmin):
end = start + n +1
diagonals.append([*range(start, end)])
start = end
for n in range(dmin, dmax):
end = start + dmin
diagonals.append([*range(start, end)])
start = end
for n in range(dmin-1, 0, -1):
end = start + n
diagonals.append([*range(start, end)])
start = end
if asnumpy:
from numpy import array
diagonals = list(map(array, diagonals))
return diagonals
def from_diagonals(diagonals, shape, dtype):
'''Returns an array constructed by the given diagonals'''
arr = np.empty(shape, dtype)
height, width = shape
for h in range(height):
row = []
for w, diag in enumerate(diagonals[h:h+width], start=h):
index = h - (w >= width) * (w - (width-1))
row.append(diag[index])
arr[h, :] = row
return arr
h, w = shape = (6,3)
arr = from_diagonals(diag_range(shape), shape, 'int')
print('Initial array:'.upper(), arr, '', sep='\n')
fliplr_arr = np.fliplr(arr)
diagonals = [fliplr_arr.diagonal(i) for i in range(-h+1, w)]
for d in diagonals:
d.flags.writeable = True # use with caution
d[:] = d[::-1]
print('Array with flipped diagonals:'.upper(), arr, sep='\n')
INITIAL ARRAY:
[[ 1 2 4]
[ 3 5 7]
[ 6 8 10]
[ 9 11 13]
[12 14 16]
[15 17 18]]
ARRAY WITH FLIPPED DIAGONALS:
[[ 1 3 6]
[ 2 5 9]
[ 4 8 12]
[ 7 11 15]
[10 14 17]
[13 16 18]]