For sake of performance, we could use a more streamlined bootstrap function.
## define custom times
t0 <- 0
t1 <- 744
## bootstrap fun
boot_fun <- function(x) {
n <- dim(x)[1]
x <- x[sample.int(n, n, replace=TRUE), ]
muhaz::muhaz(x$futime, x$fustat, min.time=t0, max.time=t1)
}
# bootstrap
set.seed(42)
R <- 999
B <- replicate(R, boot_fun(ovarian))
The results may be calculated by hand.
## extract matrix from bootstrap
r <- `colnames<-`(t(array(unlist(B[3, ]), dim=c(101, R))), B[2, ][[1]])
## calculate result
library(matrixStats) ## for fast matrix calculations
r <- cbind(x=as.numeric(colnames(r)),
y=colMeans2(r),
shape=(colMeans2(r)/colSds(r))^2,
scale=colVars(r)/colMeans2(r))
r <- cbind(r[, 1:2],
lower=qgamma(0.025, shape=r[, 'shape'] + 1, scale=r[, 'scale']),
upper=qgamma(0.975, shape=r[, 'shape'], scale=r[, 'scale']))
Gives
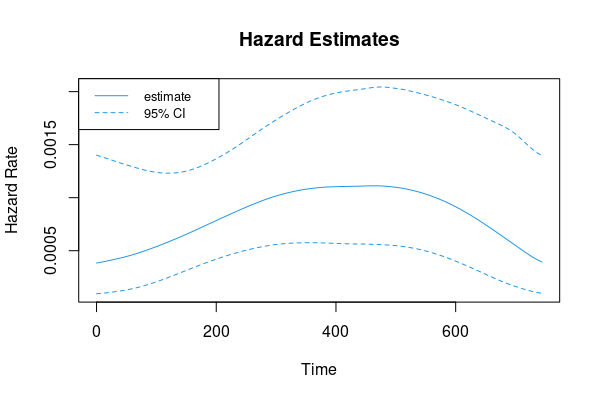
head(r)
# x y lower upper
# [1,] 0.00 0.0003836816 9.359267e-05 0.001400539
# [2,] 7.44 0.0003913992 9.746868e-05 0.001387551
# [3,] 14.88 0.0003997275 1.018897e-04 0.001374005
# [4,] 22.32 0.0004087439 1.069353e-04 0.001360212
# [5,] 29.76 0.0004178464 1.123697e-04 0.001346187
# [6,] 37.20 0.0004275531 1.184685e-04 0.001332237
range(r[, 'y'])
# [1] 0.0003836816 0.0011122910
Plot
matplot(r[, 1], r[, -1], type='l', lty=c(1, 2, 2), col=4,
xlab='Time', ylab='Hazard Rate', main='Hazard Estimates')
legend('topleft', legend=c('estimate', '95% CI'), col=4, lty=1:2, cex=.8)
![enter image description here]()
Data
data(cancer, package="survival") ## loads `ovarian` data set