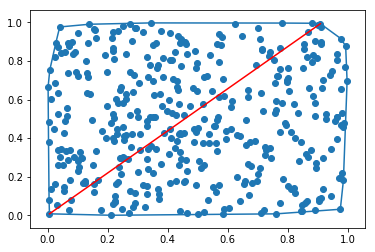
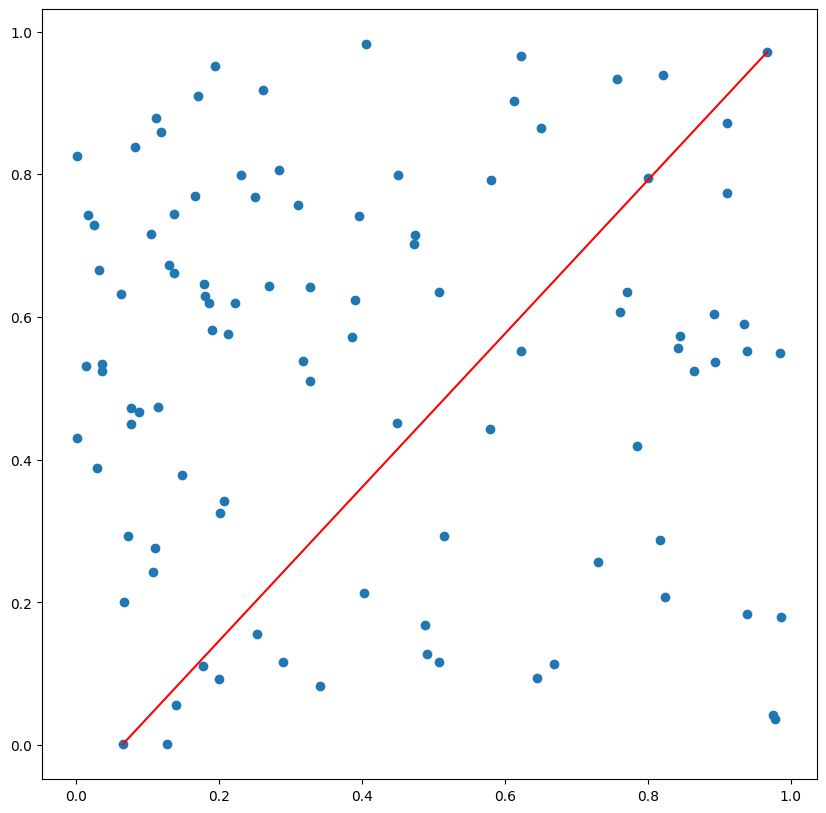
Calculating pairwise distances between all the points, selecting the two farthest points.
tl;dr - simplified example, the code:
# Standalone basic example with random data, simplified example
import numpy as np
from scipy.spatial import distance
# Generate a set of random points
pts = np.random.rand(100, 2)
distances = distance.cdist(pts, pts, 'euclidean')
maxarg = np.unravel_index(distances.argmax(), distances.shape)
print('Matrix indices of the two farthest points: %s' % (maxarg,))
print('Farthest point #1 (coords): %s' % pts[maxarg[0]])
print('Farthest point #2 (coords): %s' % pts[maxarg[1]])
Example output:
Matrix indices of the two farthest points: (11, 20)
Farthest point #1 (coords): [0.06505425 0.00118619]
Farthest point #2 (coords): [0.96760093 0.97164817]
Full example, including visualization
Code:
# Standalone basic example with random data, including visualization
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.lines import Line2D
from scipy.spatial import distance
# Generate a set of random points
pts = np.random.rand(100, 2)
distances = distance.cdist(pts, pts, 'euclidean')
maxarg = np.unravel_index(distances.argmax(), distances.shape)
print('Matrix indices of the two farthest points: %s' % (maxarg,))
print('Farthest point #1 (coords): %s' % pts[maxarg[0]])
print('Farthest point #2 (coords): %s' % pts[maxarg[1]])
# Check that the farthest distance is the same
print(distances.max())
print(distances[(maxarg)])
# Fixed size of the visualization canvas (a square)
plt.rcParams["figure.figsize"] = (10, 10)
fig = plt.figure()
ax = fig.add_subplot(111)
plt.scatter(pts.T[0], pts.T[1])
line = Line2D([pts[maxarg[0]][0], pts[maxarg[1]][0]],
[pts[maxarg[0]][1], pts[maxarg[1]][1]],
color='r')
ax.add_line(line)
plt.show()
Example output:
Matrix indices of the two farthest points: (11, 20)
Farthest point #1 (coords): [0.06505425 0.00118619]
Farthest point #2 (coords): [0.96760093 0.97164817]
1.3252875045947154
1.3252875045947154
![enter image description here]()
Why I posted this answer:
@hilberts_drinking_problem mentioned that the simple pairwise distance measure can be used, but the code he posted includes the more sophisticated Convex Hull approach. For simple problems (up to a few hundreds of points), scipy's distance matrix will be sufficient.
In the previous answer the code for the visualization was not included, and it can be very important for some use cases (to validate the result), at least it was in my case.