I am trying to plot contour lines of pressure level. I am using a netCDF file which contain the higher resolution data (ranges from 3 km to 27 km). Due to higher resolution data set, I get lot of pressure values which are not required to be plotted (rather I don't mind omitting certain contour line of insignificant values). I have written some plotting script based on the examples given in this link http://matplotlib.org/basemap/users/examples.html.
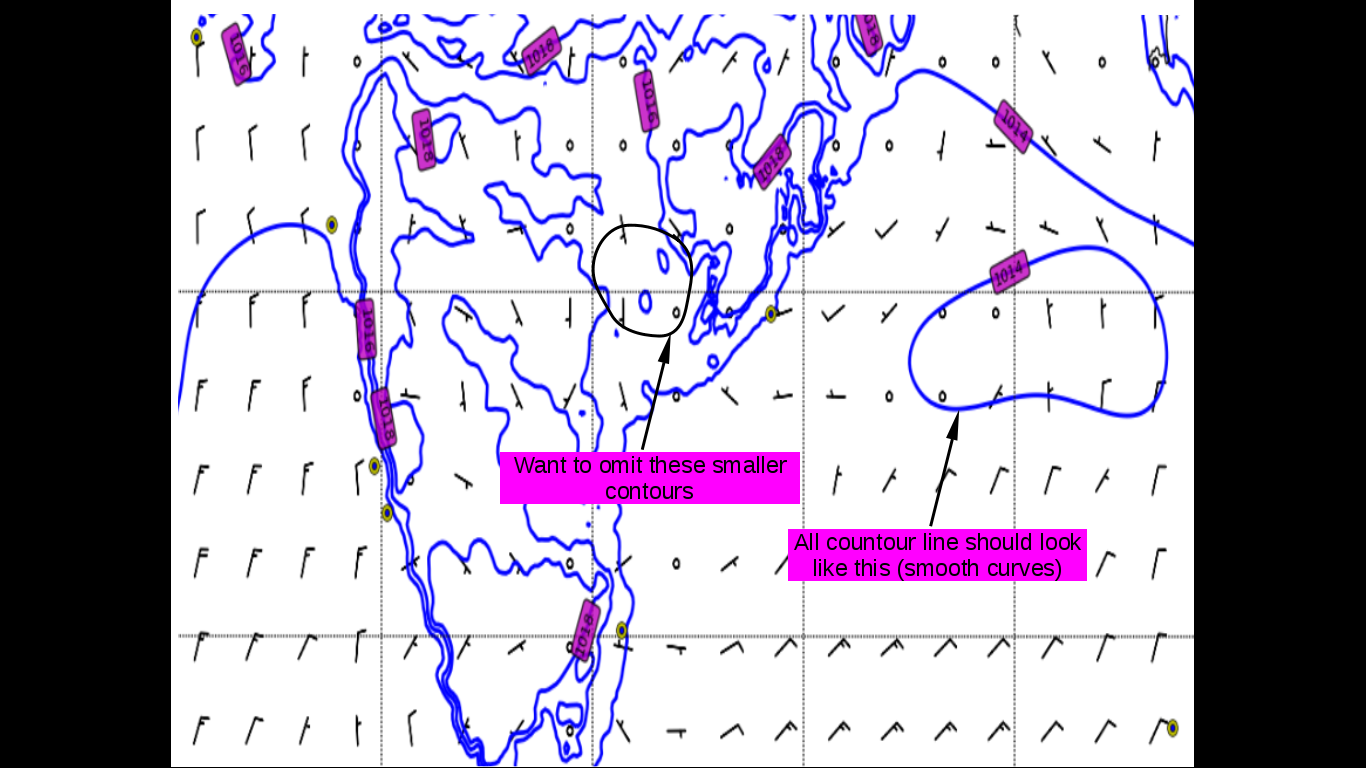
After plotting the image looks like this
From the image I have encircled the contours which are small and not required to be plotted. Also, I would like to plot all the contour lines smoother as mentioned in the above image. Overall I would like to get the contour image like this:-
Possible solution I think of are
- Find out the number of points required for plotting contour and mask/omit those lines if they are small in number.
or
- Find the area of the contour (as I want to omit only circled contour) and omit/mask those are smaller.
or
- Reduce the resolution (only contour) by increasing the distance to 50 km - 100 km.
I am able to successfully get the points using SO thread Python: find contour lines from matplotlib.pyplot.contour()
But I am not able to implement any of the suggested solution above using those points.
Any solution to implement the above suggested solution is really appreciated.
Edit:-
@ Andras Deak
I used print 'diameter is ', diameter line just above del(level.get_paths()[kp]) line to check if the code filters out the required diameter. Here is the filterd messages when I set if diameter < 15000::
diameter is 9099.66295612
diameter is 13264.7838257
diameter is 445.574234531
diameter is 1618.74618114
diameter is 1512.58974168
However the resulting image does not have any effect. All look same as posed image above. I am pretty sure that I have saved the figure (after plotting the wind barbs).
Regarding the solution for reducing the resolution, plt.contour(x[::2,::2],y[::2,::2],mslp[::2,::2]) it works. I have to apply some filter to make the curve smooth.
Full working example code for removing lines:-
Here is the example code for your review
#!/usr/bin/env python
from netCDF4 import Dataset
import matplotlib
matplotlib.use('agg')
import matplotlib.pyplot as plt
import numpy as np
import scipy.ndimage
from mpl_toolkits.basemap import interp
from mpl_toolkits.basemap import Basemap
# Set default map
west_lon = 68
east_lon = 93
south_lat = 7
north_lat = 23
nc = Dataset('ncfile.nc')
# Get this variable for later calucation
temps = nc.variables['T2']
time = 0 # We will take only first interval for this example
# Draw basemap
m = Basemap(projection='merc', llcrnrlat=south_lat, urcrnrlat=north_lat,
llcrnrlon=west_lon, urcrnrlon=east_lon, resolution='l')
m.drawcoastlines()
m.drawcountries(linewidth=1.0)
# This sets the standard grid point structure at full resolution
x, y = m(nc.variables['XLONG'][0], nc.variables['XLAT'][0])
# Set figure margins
width = 10
height = 8
plt.figure(figsize=(width, height))
plt.rc("figure.subplot", left=.001)
plt.rc("figure.subplot", right=.999)
plt.rc("figure.subplot", bottom=.001)
plt.rc("figure.subplot", top=.999)
plt.figure(figsize=(width, height), frameon=False)
# Convert Surface Pressure to Mean Sea Level Pressure
stemps = temps[time] + 6.5 * nc.variables['HGT'][time] / 1000.
mslp = nc.variables['PSFC'][time] * np.exp(9.81 / (287.0 * stemps) * nc.variables['HGT'][time]) * 0.01 + (
6.7 * nc.variables['HGT'][time] / 1000)
# Contour only at 2 hpa interval
level = []
for i in range(mslp.min(), mslp.max(), 1):
if i % 2 == 0:
if i >= 1006 and i <= 1018:
level.append(i)
# Save mslp values to upload to SO thread
# np.savetxt('mslp.txt', mslp, fmt='%.14f', delimiter=',')
P = plt.contour(x, y, mslp, V=2, colors='b', linewidths=2, levels=level)
# Solution suggested by Andras Deak
for level in P.collections:
for kp,path in enumerate(level.get_paths()):
# include test for "smallness" of your choice here:
# I'm using a simple estimation for the diameter based on the
# x and y diameter...
verts = path.vertices # (N,2)-shape array of contour line coordinates
diameter = np.max(verts.max(axis=0) - verts.min(axis=0))
if diameter < 15000: # threshold to be refined for your actual dimensions!
#print 'diameter is ', diameter
del(level.get_paths()[kp]) # no remove() for Path objects:(
#level.remove() # This does not work. produces ValueError: list.remove(x): x not in list
plt.gcf().canvas.draw()
plt.savefig('dummy', bbox_inches='tight')
plt.close()
After the plot is saved I get the same image
You can see that the lines are not removed yet. Here is the link to mslp array which we are trying to play with http://www.mediafire.com/download/7vi0mxqoe0y6pm9/mslp.txt
If you want x and y data which are being used in the above code, I can upload for your review.
Smooth line
You code to remove the smaller circles working perfectly. However the other question I have asked in the original post (smooth line) does not seems to work. I have used your code to slice the array to get minimal values and contoured it. I have used the following code to reduce the array size:-
slice = 15
CS = plt.contour(x[::slice,::slice],y[::slice,::slice],mslp[::slice,::slice], colors='b', linewidths=1, levels=levels)
The result is below.
After searching for few hours I found this SO thread having simmilar issue:-
Regridding regular netcdf data
But none of the solution provided over there works.The questions similar to mine above does not have proper solutions. If this issue is solved then the code is perfect and complete.

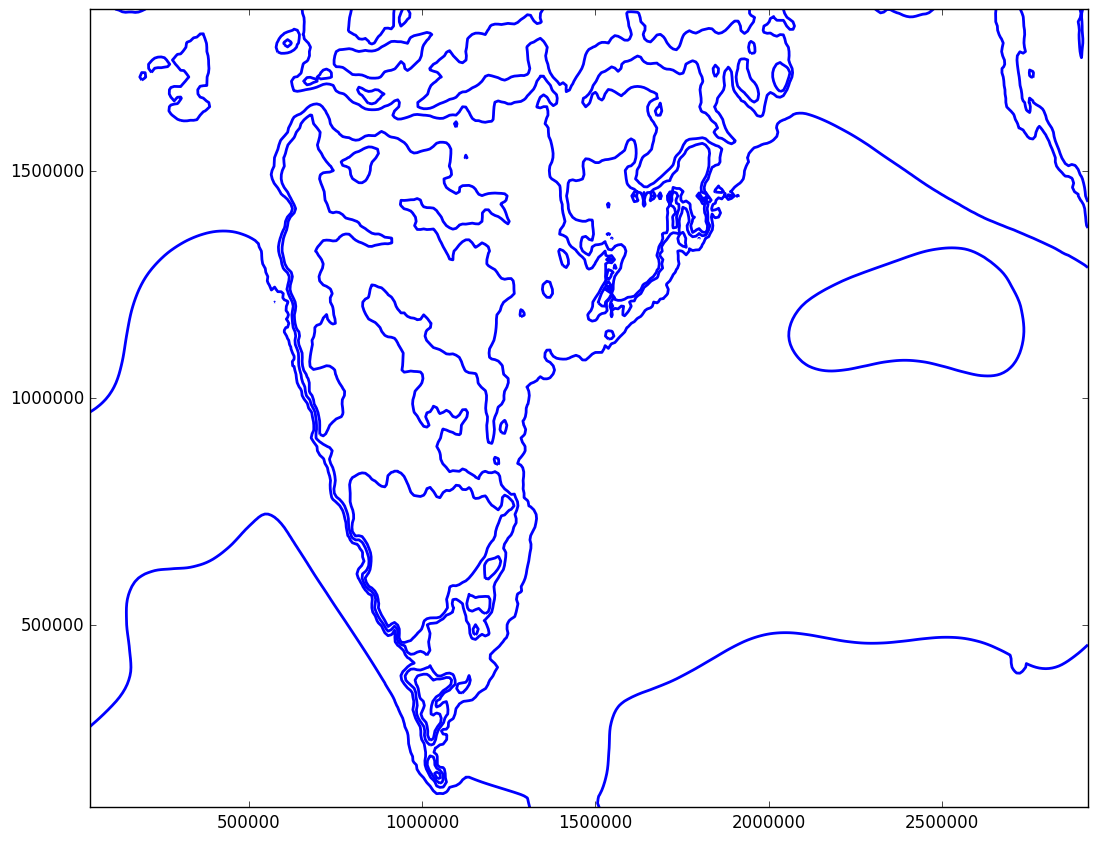
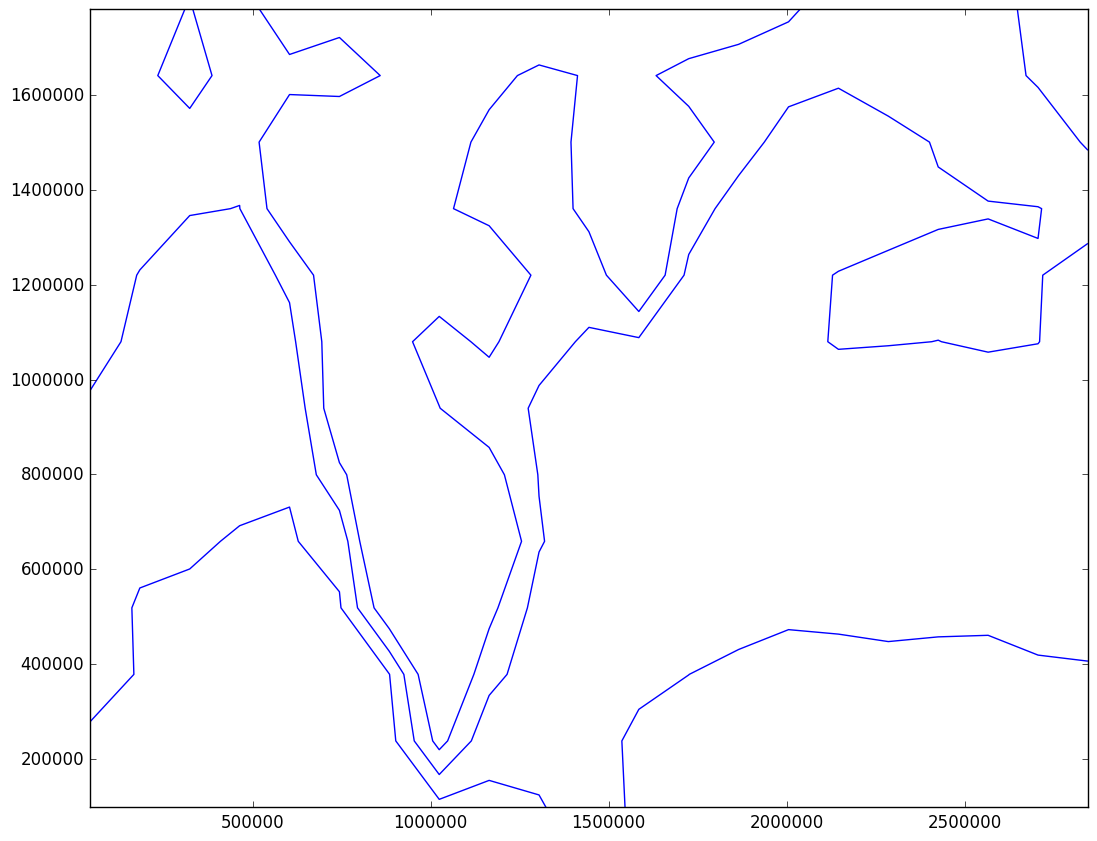






plt.gcf().canvas.draw()the saved fig looks same. I used this codeP = plt.contour(x, y, mslp, V=2, colors='b', linewidths=2, levels=level)for plotting the image. How do I update the image which are saved as .png file? Also, you said thatdecreasing the resolution of your contour()and I have been trying to do this for past two days without success. Please show some code for reducing the resolution (it is to plotted on basemap) and I will accept the answer. – Hussite