it doesn't look like you'll get anything worthwhile following that procedure, there are much better techniques for handling unexpected data. googling for "outlier detection" would be a good start.
with that said, here's how to answer your question:
start by pulling in libraries and getting some data:
import matplotlib.pyplot as plt
import numpy as np
Y = np.array([
0.00441025, 0.0049001 , 0.01041189, 0.47368389, 0.34841961,
0.3487533 , 0.35067096, 0.31142986, 0.3268407 , 0.38099566,
0.3933048 , 0.3479948 , 0.02359819, 0.36329588, 0.42535543,
0.01308297, 0.53873956, 0.6511364 , 0.61865282, 0.64750302,
0.6630047 , 0.66744816, 0.71759617, 0.05965622, 0.71335208,
0.71992683, 0.61635697, 0.12985441, 0.73410642, 0.77318621,
0.75675988, 0.03003641, 0.77527201, 0.78673995, 0.05049178,
0.55139476, 0.02665514, 0.61664748, 0.81121749, 0.05521697,
0.63404375, 0.32649395, 0.36828268, 0.68981099, 0.02874863,
0.61574739])
X = np.linspace(0, 1, len(Y))
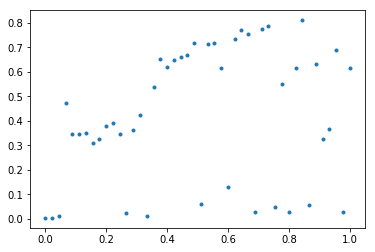
next do an initial plot of the data:
plt.plot(X, Y, '.')
![initial data plot]()
as this lets you see what we're dealing with and whether a polynomial would ever be a good fit --- short answer is that this method isn't going to get very far with this sort of data
at this point we should stop, but to answer the question I'll go on, mostly following your polynomial fitting code:
poly_degree = 5
sd_cutoff = 1 # 2 keeps everything
coeffs = np.polyfit(X, Y, poly_degree)
poly_eqn = np.poly1d(coeffs)
Y_hat = poly_eqn(X)
delta = Y - Y_hat
sd_p = np.std(delta)
ok = abs(delta) < sd_p * sd_cutoff
hopefully this makes sense, I use a higher degree polynomial and only cutoff at 1SD because otherwise nothing will be thrown away. the ok array contains True values for those points that are within sd_cutoff standard deviations
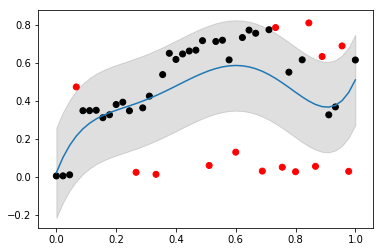
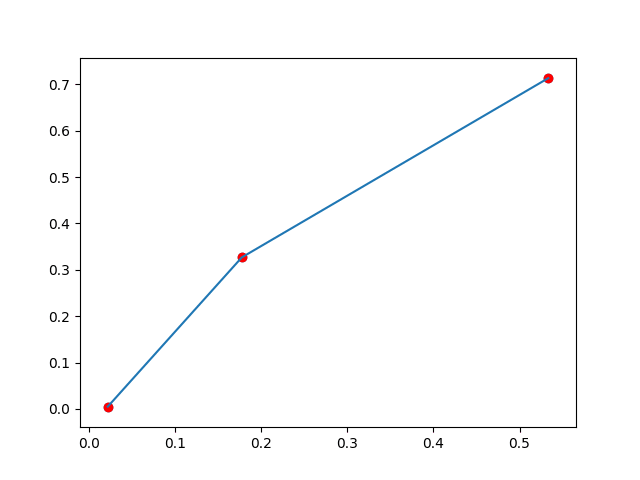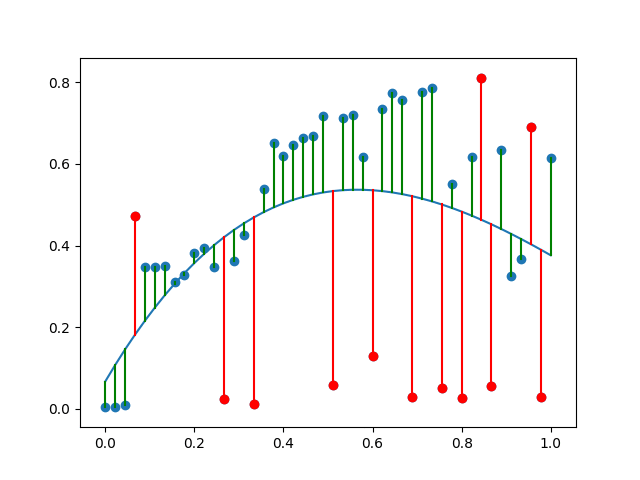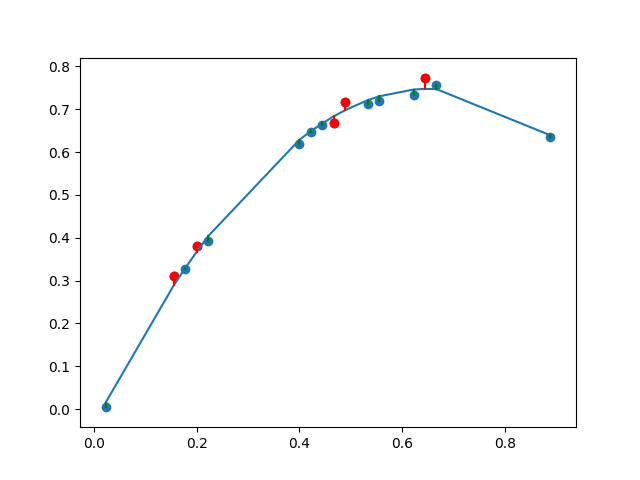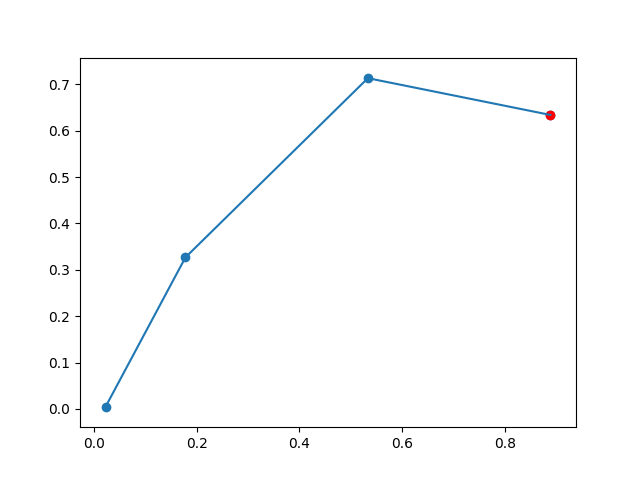
to check this, I'd then do another plot. something like:
plt.scatter(X, Y, color=np.where(ok, 'k', 'r'))
plt.fill_between(
X,
Y_hat - sd_p * sd_cutoff,
Y_hat + sd_p * sd_cutoff,
color='#00000020')
plt.plot(X, Y_hat)
which gives me:
![data with poly and 1sd]()
so the black dots are the points to keep (i.e. X[ok] gives me these back, and np.where(ok) gives you indicies).
you can play around with the parameters, but you probably want a distribution with fatter tails (e.g. a Student's T-distribution) but, as I said above, using Google for outlier detection would be my suggestion

mean of curve. – Motivation