The purpose of this mathematical function is to compute a distance between two (or more) protein structures using dihedral angles:
It is very useful in structural biology, for example. And I already code this function in python using numpy, but the goal is to have a faster implementation. As computation time reference, I use the euclidean distance function available in the scikit-learn package.
Here the code I have for the moment:
import numpy as np
import numexpr as ne
from sklearn.metrics.pairwise import euclidean_distances
# We have 10000 structures with 100 dihedral angles
n = 10000
m = 100
# Generate some random data
c = np.random.rand(n,m)
# Generate random int number
x = np.random.randint(c.shape[0])
print c.shape, x
# First version with numpy of the dihedral_distances function
def dihedral_distances(a, b):
l = 1./a.shape[0]
return np.sqrt(l* np.sum((0.5)*(1. - np.cos(a-b)), axis=1))
# Accelerated version with numexpr
def dihedral_distances_ne(a, b):
l = 1./a.shape[0]
tmp = ne.evaluate('sum((0.5)*(1. - cos(a-b)), axis=1)')
return ne.evaluate('sqrt(l* tmp)')
# The function of reference I try to be close as possible
# in term of computation time
%timeit euclidean_distances(c[x,:], c)[0]
1000 loops, best of 3: 1.07 ms per loop
# Computation time of the first version of the dihedral_distances function
# We choose randomly 1 structure among the 10000 structures.
# And we compute the dihedral distance between this one and the others
%timeit dihedral_distances(c[x,:], c)
10 loops, best of 3: 21.5 ms per loop
# Computation time of the accelerated function with numexpr
%timeit dihedral_distances_ne(c[x,:], c)
100 loops, best of 3: 9.44 ms per loop
9.44 ms it's very fast, but it's very slow if you need to run it a million times. Now the question is, how to do that? What is the next step? Cython? PyOpenCL? I have some experience with PyOpenCL, however I never code something as elaborate as this one. I don't know if it's possible to compute the dihedral distances in one step on GPU as I do with numpy and how to proceed.
Thank you for helping me!
EDIT: Thank you guys! I am currently working on the full solution and once it's finished I will put the code here.
CYTHON VERSION:
%load_ext cython
import numpy as np
np.random.seed(1234)
n = 10000
m = 100
c = np.random.rand(n,m)
x = np.random.randint(c.shape[0])
print c.shape, x
%%cython --compile-args=-fopenmp --link-args=-fopenmp --force
import numpy as np
cimport numpy as np
from libc.math cimport sqrt, cos
cimport cython
from cython.parallel cimport parallel, prange
# Define a function pointer to a metric
ctypedef double (*metric)(double[: ,::1], np.intp_t, np.intp_t)
cdef extern from "math.h" nogil:
double cos(double x)
double sqrt(double x)
@cython.boundscheck(False)
@cython.wraparound(False)
@cython.cdivision(True)
cdef double dihedral_distances(double[:, ::1] a, np.intp_t i1, np.intp_t i2):
cdef double res
cdef int m
cdef int j
res = 0.
m = a.shape[1]
for j in range(m):
res += 1. - cos(a[i1, j] - a[i2, j])
res /= 2.*m
return sqrt(res)
@cython.boundscheck(False)
@cython.wraparound(False)
@cython.cdivision(True)
cdef double dihedral_distances_p(double[:, ::1] a, np.intp_t i1, np.intp_t i2):
cdef double res
cdef int m
cdef int j
res = 0.
m = a.shape[1]
with nogil, parallel(num_threads=2):
for j in prange(m, schedule='dynamic'):
res += 1. - cos(a[i1, j] - a[i2, j])
res /= 2.*m
return sqrt(res)
@cython.boundscheck(False)
@cython.wraparound(False)
def pairwise(double[: ,::1] c not None, np.intp_t x, p = True):
cdef metric dist_func
if p:
dist_func = &dihedral_distances_p
else:
dist_func = &dihedral_distances
cdef np.intp_t i, n_structures
n_samples = c.shape[0]
cdef double[::1] res = np.empty(n_samples)
for i in range(n_samples):
res[i] = dist_func(c, x, i)
return res
%timeit pairwise(c, x, False)
100 loops, best of 3: 17 ms per loop
# Parallel version
%timeit pairwise(c, x, True)
10 loops, best of 3: 37.1 ms per loop
So I follow your link to create the cython version of the dihedral distances function. We gain some speed, not so much, but it is still slower than the numexpr version (17ms vs 9.44ms). So I tried to parallelize the function using prange and it is worse (37.1ms vs 17ms vs 9.4ms)!
Do I miss something?

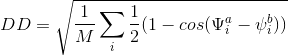
*0.5outside of the sum; and, 2) sum thecosbefore subtracting the from1(which will be more accurate as well, since the sum will hang around 1). For me these bring it from 25ms to 17ms. I know you're looking for more, but this is what I got and thought it might help a bit. – AntediluvianRuntimeWarning: invalid value encountered in sqrt. – Gabisqrt(sum(1 - cos(x)))becomessqrt(N - sum(cos(x))). Did you remember the N? – Antediluvian