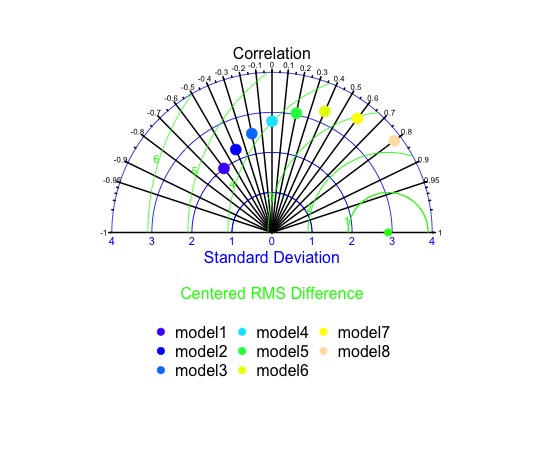
One option is to use a different package, such as openair, which might be more flexible. Because of your specific requirements it might be easier to use code designed for your requirements. I wrote some code to generate the following plot, which is close to your desired plot. You can hack the code to adjust the plot to your desired format.
![enter image description here]()
# code to make a Taylor diagram
# formulas found in http://www-pcmdi.llnl.gov/about/staff/Taylor/CV/Taylor_diagram_primer.pdf
# and http://rainbow.llnl.gov/publications/pdf/55.pdf
# correlations and tick marks (only major will have a line to the center)
# minor will have a tick mark
correlation_major <- c(seq(-1,1,0.1),-0.95,0.95)
correlation_minor <- c(seq(-1,-0.95,0.01),seq(-0.9,9,0.05),seq(0.95,1,0.01))
# test standard deviation tick marks (only major will have a line)
sigma_test_major <- seq(1,4,1)
sigma_test_minor <- seq(0.5,4,0.5)
# rms lines locations
rms_major <- seq(1,6,1)
# reference standard deviation (observed)
sigma_reference <- 2.9
# color schemes for the liens
correlation_color <- 'black'
sigma_test_color <- 'blue'
rms_color <- 'green'
# line types
correlation_type <- 1
sigma_test_type <- 1
rms_type <- 1
# plot parameters
par(pty='s')
par(mar=c(3,3,3,3)+0.1)
# creating plot with correct space based on the sigma_test limits
plot(NA
,NA
,xlim=c(-1*max(sigma_test_major),max(sigma_test_major))
,ylim=c(-1*max(sigma_test_major),max(sigma_test_major))
,xaxt='n'
,yaxt='n'
,xlab=''
,ylab=''
,bty='n')
#### adding sigma_test (standard deviation)
# adding semicircles
for(i in 1:length(sigma_test_major)){
lines(sigma_test_major[i]*cos(seq(0,pi,pi/1000))
,sigma_test_major[i]*sin(seq(0,pi,pi/1000))
,col=sigma_test_color
,lty=sigma_test_type
,lwd=1
)
}
# adding horizontal axis
lines(c(-1*max(sigma_test_major),max(sigma_test_major))
,c(0,0)
,col=sigma_test_color
,lty=sigma_test_type
,lwd=1)
# adding labels
text(c(-1*sigma_test_major,0,sigma_test_major)
,-0.2
,as.character(c(-1*sigma_test_major,0,sigma_test_major))
,col=sigma_test_color
,cex=0.7)
# adding title
text(0
,-0.6
,"Standard Deviation"
,col=sigma_test_color
,cex=1)
#### adding correlation lines, tick marks, and lables
# adding lines
for(i in 1:length(correlation_major)){
lines(c(0,1.02*max(sigma_test_major)*cos(acos(correlation_major[i])))
,c(0,1.02*max(sigma_test_major)*sin(acos(correlation_major[i])))
,lwd=2
,lty=correlation_type
,col=correlation_color
)
}
# adding minor tick marks for correlation
for(i in 1:length(correlation_minor)){
lines(max(sigma_test_major)*cos(acos(correlation_minor[i]))*c(1,1.01)
,max(sigma_test_major)*sin(acos(correlation_minor[i]))*c(1,1.01)
,lwd=2
,lty=correlation_type
,col=correlation_color
)
}
# adding labels for correlation
text(1.05*max(sigma_test_major)*cos(acos(correlation_major))
,1.05*max(sigma_test_major)*sin(acos(correlation_major))
,as.character(correlation_major)
,col=correlation_color
,cex=0.5)
# adding correlation title
text(0
,max(sigma_test_major)+0.5
,"Correlation"
,col=correlation_color
,cex=1)
#### adding rms difference lines
# adding rms semicircles
for(i in 1:length(rms_major)){
inds <- which((rms_major[i]*cos(seq(0,pi,pi/1000))+sigma_reference)^2 + (rms_major[i]*sin(seq(0,pi,pi/1000)))^2 < max(sigma_test_major)^2)
lines(rms_major[i]*cos(seq(0,pi,pi/1000))[inds]+sigma_reference
,rms_major[i]*sin(seq(0,pi,pi/1000))[inds]
,col=rms_color
,lty=rms_type
,lwd=1
)
}
# adding observed point
points(sigma_reference
,0
,pch=19
,col=rms_color
,cex=1)
# adding labels for the rms lines
text(-1*rms_major*cos(pi*rms_major/40)+sigma_reference
, rms_major*sin(pi*rms_major/40)
,as.character(rms_major)
,col=rms_color
,cex=0.7
,adj=1)
# adding title
text(0
,-1.5
,'Centered RMS Difference'
,col=rms_color
,cex=1
,adj=0.5)
###################### adding points #####################
names <- paste("model",seq(1,8),sep='')
correl_names <- seq(-0.6,0.8,by=0.2)
std_names <- seq(2,4,by=0.26)
color_names <- topo.colors(length(names))
points(std_names*cos(acos(correl_names))
,std_names*sin(acos(correl_names))
,col=color_names
,pch=19
,cex=1.5)
# making legend
par(xpd=TRUE)
legend(0,-2
,names
,pc=19
,col=color_names
,ncol=3
,bty='n'
,xjust=0.5)