I have a working implementation of multivariable linear regression using gradient descent in R. I'd like to see if I can use what I have to run a stochastic gradient descent. I'm not sure if this is really inefficient or not. For example, for each value of α I want to perform 500 SGD iterations and be able to specify the number of randomly picked samples in each iteration. It would be nice to do this so I could see how the number of samples influences the results. I'm having trouble through with the mini-batching and I want to be able to easily plot the results.
This is what I have so far:
# Read and process the datasets
# download the files from GitHub
download.file("https://raw.githubusercontent.com/dbouquin/IS_605/master/sgd_ex_data/ex3x.dat", "ex3x.dat", method="curl")
x <- read.table('ex3x.dat')
# we can standardize the x vaules using scale()
x <- scale(x)
download.file("https://raw.githubusercontent.com/dbouquin/IS_605/master/sgd_ex_data/ex3y.dat", "ex3y.dat", method="curl")
y <- read.table('ex3y.dat')
# combine the datasets
data3 <- cbind(x,y)
colnames(data3) <- c("area_sqft", "bedrooms","price")
str(data3)
head(data3)
################ Regular Gradient Descent
# http://www.r-bloggers.com/linear-regression-by-gradient-descent/
# vector populated with 1s for the intercept coefficient
x1 <- rep(1, length(data3$area_sqft))
# appends to dfs
# create x-matrix of independent variables
x <- as.matrix(cbind(x1,x))
# create y-matrix of dependent variables
y <- as.matrix(y)
L <- length(y)
# cost gradient function: independent variables and values of thetas
cost <- function(x,y,theta){
gradient <- (1/L)* (t(x) %*% ((x%*%t(theta)) - y))
return(t(gradient))
}
# GD simultaneous update algorithm
# https://www.coursera.org/learn/machine-learning/lecture/8SpIM/gradient-descent
GD <- function(x, alpha){
theta <- matrix(c(0,0,0), nrow=1)
for (i in 1:500) {
theta <- theta - alpha*cost(x,y,theta)
theta_r <- rbind(theta_r,theta)
}
return(theta_r)
}
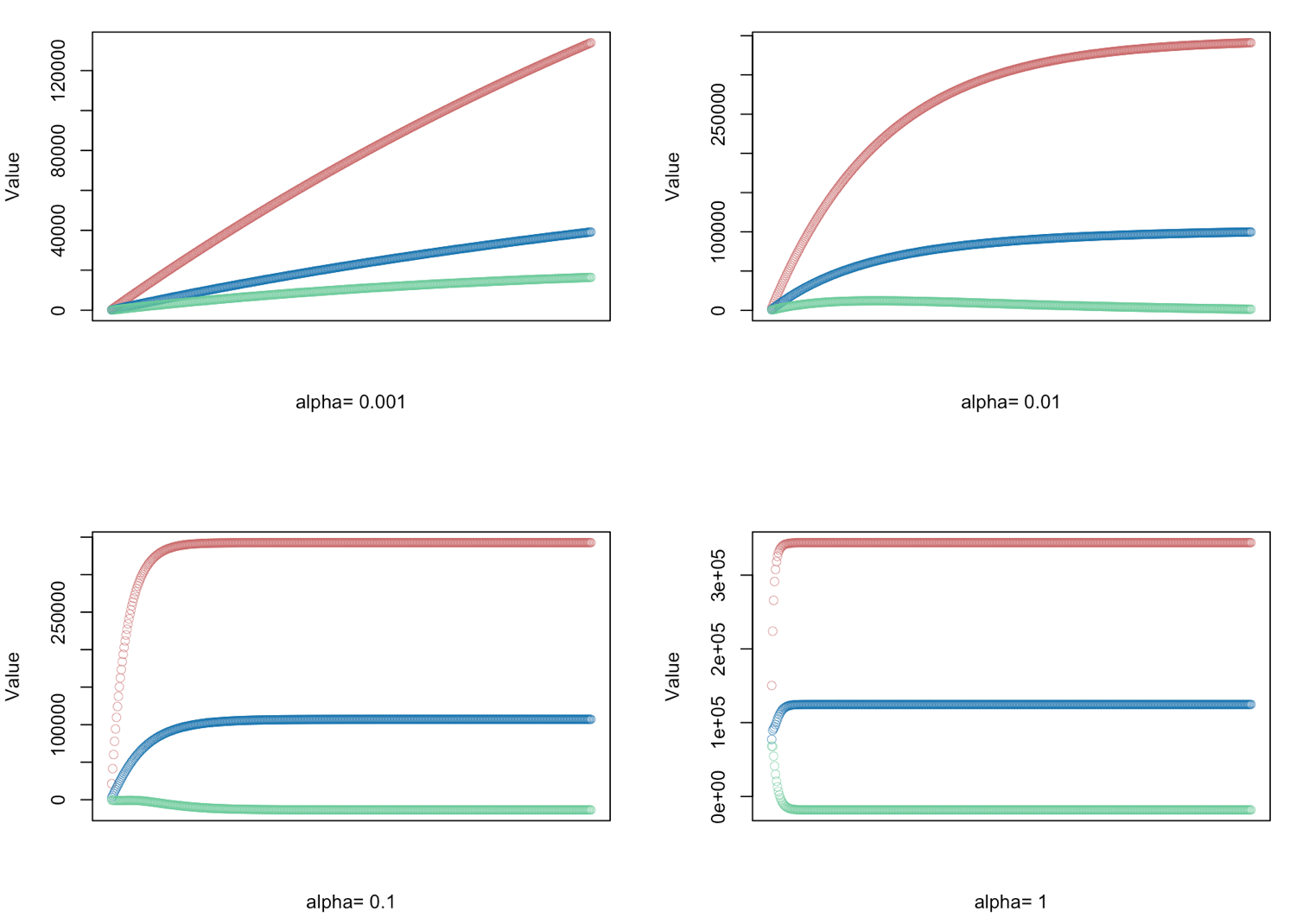
# gradient descent α = (0.001, 0.01, 0.1, 1.0) - defined for 500 iterations
alphas <- c(0.001,0.01,0.1,1.0)
# Plot price, area in square feet, and the number of bedrooms
# create empty vector theta_r
theta_r<-c()
for(i in 1:length(alphas)) {
result <- GD(x, alphas[i])
# red = price
# blue = sq ft
# green = bedrooms
plot(result[,1],ylim=c(min(result),max(result)),col="#CC6666",ylab="Value",lwd=0.35,
xlab=paste("alpha=", alphas[i]),xaxt="n") #suppress auto x-axis title
lines(result[,2],type="b",col="#0072B2",lwd=0.35)
lines(result[,3],type="b",col="#66CC99",lwd=0.35)
}
Is it more practical to find a way to use sgd()? I can't seem to figure out how to have the level of control I'm looking for with the sgd package

sgdhas two arguments,model.controlandsgd.controlboth of which have a pretty big list of options you can pass. do you want more control than that? what else are you trying to do? – Splitlevelsample500 of your total and use the subset of data in your models. remember toset.seed– Splitlevel?sgdhas a multivariable linear example although it is quite simple. there is also the vignette – Splitlevelmodel.controlandsgd.controlappear to be controlled bysgd:::valid_model_controlandsgd:::valid_sgd_control, although I don't see options for the number of observations. Given that sgd is guaranteed to be optimal with batch size == 1, there may not be an option. Typically batch size is only specified to control the learning time (in computation time, not number of iterations)... Since the package is under active development, I suggest you raise an issue with the authors... even if you use rawr's wrapper below – Geomancer