How do I initialize weights and biases of a network (via e.g. He or Xavier initialization)?
Single layer
To initialize the weights of a single layer, use a function from torch.nn.init. For instance:
conv1 = torch.nn.Conv2d(...)
torch.nn.init.xavier_uniform(conv1.weight)
Alternatively, you can modify the parameters by writing to conv1.weight.data (which is a torch.Tensor). Example:
conv1.weight.data.fill_(0.01)
The same applies for biases:
conv1.bias.data.fill_(0.01)
nn.Sequential or custom nn.Module
Pass an initialization function to torch.nn.Module.apply. It will initialize the weights in the entire nn.Module recursively.
apply(fn): Applies
fnrecursively to every submodule (as returned by.children()) as well as self. Typical use includes initializing the parameters of a model (see also torch-nn-init).
Example:
def init_weights(m):
if isinstance(m, nn.Linear):
torch.nn.init.xavier_uniform(m.weight)
m.bias.data.fill_(0.01)
net = nn.Sequential(nn.Linear(2, 2), nn.Linear(2, 2))
net.apply(init_weights)
reset_parameters method in the source code of many modules. Should I override the method for weight initialization? –
Spoilage nn.init.xavier_uniform is now deprecated in favor of nn.init.xavier_uniform_ –
Hough Linear layer is init.kaiming_uniform_(self.weight, a=math.sqrt(5)). –
Lunarian We compare different mode of weight-initialization using the same neural-network(NN) architecture.
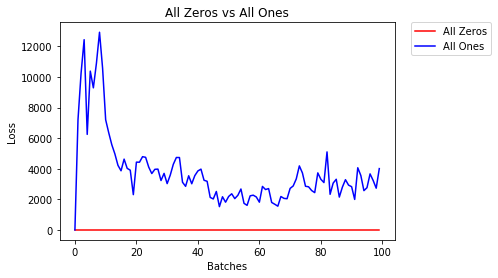
All Zeros or Ones
If you follow the principle of Occam's razor, you might think setting all the weights to 0 or 1 would be the best solution. This is not the case.
With every weight the same, all the neurons at each layer are producing the same output. This makes it hard to decide which weights to adjust.
# initialize two NN's with 0 and 1 constant weights
model_0 = Net(constant_weight=0)
model_1 = Net(constant_weight=1)
- After 2 epochs:
Validation Accuracy
9.625% -- All Zeros
10.050% -- All Ones
Training Loss
2.304 -- All Zeros
1552.281 -- All Ones
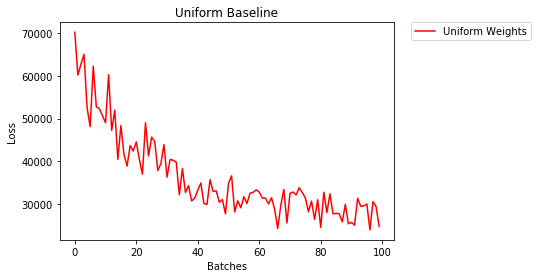
Uniform Initialization
A uniform distribution has the equal probability of picking any number from a set of numbers.
Let's see how well the neural network trains using a uniform weight initialization, where low=0.0 and high=1.0.
Below, we'll see another way (besides in the Net class code) to initialize the weights of a network. To define weights outside of the model definition, we can:
- Define a function that assigns weights by the type of network layer, then
- Apply those weights to an initialized model using
model.apply(fn), which applies a function to each model layer.
# takes in a module and applies the specified weight initialization
def weights_init_uniform(m):
classname = m.__class__.__name__
# for every Linear layer in a model..
if classname.find('Linear') != -1:
# apply a uniform distribution to the weights and a bias=0
m.weight.data.uniform_(0.0, 1.0)
m.bias.data.fill_(0)
model_uniform = Net()
model_uniform.apply(weights_init_uniform)
- After 2 epochs:
Validation Accuracy
36.667% -- Uniform Weights
Training Loss
3.208 -- Uniform Weights
General rule for setting weights
The general rule for setting the weights in a neural network is to set them to be close to zero without being too small.
Good practice is to start your weights in the range of [-y, y] where
y=1/sqrt(n)
(n is the number of inputs to a given neuron).
# takes in a module and applies the specified weight initialization
def weights_init_uniform_rule(m):
classname = m.__class__.__name__
# for every Linear layer in a model..
if classname.find('Linear') != -1:
# get the number of the inputs
n = m.in_features
y = 1.0/np.sqrt(n)
m.weight.data.uniform_(-y, y)
m.bias.data.fill_(0)
# create a new model with these weights
model_rule = Net()
model_rule.apply(weights_init_uniform_rule)
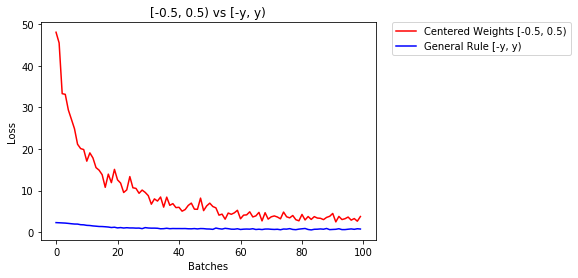
below we compare performance of NN, weights initialized with uniform distribution [-0.5,0.5) versus the one whose weight is initialized using general rule
- After 2 epochs:
Validation Accuracy
75.817% -- Centered Weights [-0.5, 0.5)
85.208% -- General Rule [-y, y)
Training Loss
0.705 -- Centered Weights [-0.5, 0.5)
0.469 -- General Rule [-y, y)
normal distribution to initialize the weights
The normal distribution should have a mean of 0 and a standard deviation of
y=1/sqrt(n), where n is the number of inputs to NN
## takes in a module and applies the specified weight initialization
def weights_init_normal(m):
'''Takes in a module and initializes all linear layers with weight
values taken from a normal distribution.'''
classname = m.__class__.__name__
# for every Linear layer in a model
if classname.find('Linear') != -1:
y = m.in_features
# m.weight.data shoud be taken from a normal distribution
m.weight.data.normal_(0.0,1/np.sqrt(y))
# m.bias.data should be 0
m.bias.data.fill_(0)
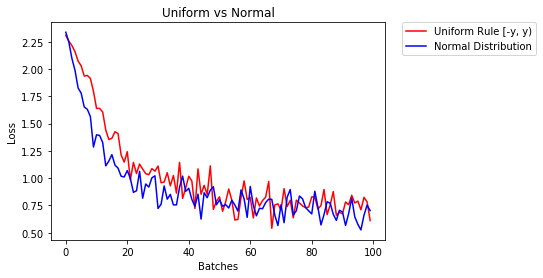
below we show the performance of two NN one initialized using uniform-distribution and the other using normal-distribution
- After 2 epochs:
Validation Accuracy
85.775% -- Uniform Rule [-y, y)
84.717% -- Normal Distribution
Training Loss
0.329 -- Uniform Rule [-y, y)
0.443 -- Normal Distribution
To initialize layers, you typically don't need to do anything.
PyTorch will do it for you. If you think about it, this makes a lot of sense. Why should we initialize layers, when PyTorch can do that following the latest trends?
For instance, the Linear layer's __init__ method will do Kaiming He initialization:
init.kaiming_uniform_(self.weight, a=math.sqrt(5))
if self.bias is not None:
fan_in, _ = init._calculate_fan_in_and_fan_out(self.weight)
bound = 1 / math.sqrt(fan_in) if fan_in > 0 else 0
init.uniform_(self.bias, -bound, bound)
Similarly, this holds for other layers types. For e.g., Conv2d, check here.
NOTE: The advantage of proper initialization is faster training speed. If your problem requires special initialization, you can still do it afterwards.
xavier_uniform initialization for the weights (with biases initialized to 0), rather than using the default initialization, my validation accuracy after 30 epochs of RMSprop increased from 82% to 86%. I also got 86% validation accuracy when using Pytorch's built-in VGG16 model (not pre-trained), so I think I implemented it correctly. (I used a learning rate of 0.00001.) –
Holder import torch.nn as nn
# a simple network
rand_net = nn.Sequential(nn.Linear(in_features, h_size),
nn.BatchNorm1d(h_size),
nn.ReLU(),
nn.Linear(h_size, h_size),
nn.BatchNorm1d(h_size),
nn.ReLU(),
nn.Linear(h_size, 1),
nn.ReLU())
# initialization function, first checks the module type,
# then applies the desired changes to the weights
def init_normal(m):
if type(m) == nn.Linear:
nn.init.uniform_(m.weight)
# use the modules apply function to recursively apply the initialization
rand_net.apply(init_normal)
If you want some extra flexibility, you can also set the weights manually.
Say you have input of all ones:
import torch
import torch.nn as nn
input = torch.ones((8, 8))
print(input)
tensor([[1., 1., 1., 1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1., 1., 1., 1.],
[1., 1., 1., 1., 1., 1., 1., 1.]])
And you want to make a dense layer with no bias (so we can visualize):
d = nn.Linear(8, 8, bias=False)
Set all the weights to 0.5 (or anything else):
d.weight.data = torch.full((8, 8), 0.5)
print(d.weight.data)
The weights:
Out[14]:
tensor([[0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000],
[0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000],
[0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000],
[0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000],
[0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000],
[0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000],
[0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000],
[0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000, 0.5000]])
All your weights are now 0.5. Pass the data through:
d(input)
Out[13]:
tensor([[4., 4., 4., 4., 4., 4., 4., 4.],
[4., 4., 4., 4., 4., 4., 4., 4.],
[4., 4., 4., 4., 4., 4., 4., 4.],
[4., 4., 4., 4., 4., 4., 4., 4.],
[4., 4., 4., 4., 4., 4., 4., 4.],
[4., 4., 4., 4., 4., 4., 4., 4.],
[4., 4., 4., 4., 4., 4., 4., 4.],
[4., 4., 4., 4., 4., 4., 4., 4.]], grad_fn=<MmBackward>)
Remember that each neuron receives 8 inputs, all of which have weight 0.5 and value of 1 (and no bias), so it sums up to 4 for each.
Sorry for being so late, I hope my answer will help.
To initialise weights with a normal distribution use:
torch.nn.init.normal_(tensor, mean=0, std=1)
Or to use a constant distribution write:
torch.nn.init.constant_(tensor, value)
Or to use an uniform distribution:
torch.nn.init.uniform_(tensor, a=0, b=1) # a: lower_bound, b: upper_bound
You can check other methods to initialise tensors here
Iterate over parameters
If you cannot use apply for instance if the model does not implement Sequential directly:
Same for all
# see UNet at https://github.com/milesial/Pytorch-UNet/tree/master/unet
def init_all(model, init_func, *params, **kwargs):
for p in model.parameters():
init_func(p, *params, **kwargs)
model = UNet(3, 10)
init_all(model, torch.nn.init.normal_, mean=0., std=1)
# or
init_all(model, torch.nn.init.constant_, 1.)
Depending on shape
def init_all(model, init_funcs):
for p in model.parameters():
init_func = init_funcs.get(len(p.shape), init_funcs["default"])
init_func(p)
model = UNet(3, 10)
init_funcs = {
1: lambda x: torch.nn.init.normal_(x, mean=0., std=1.), # can be bias
2: lambda x: torch.nn.init.xavier_normal_(x, gain=1.), # can be weight
3: lambda x: torch.nn.init.xavier_uniform_(x, gain=1.), # can be conv1D filter
4: lambda x: torch.nn.init.xavier_uniform_(x, gain=1.), # can be conv2D filter
"default": lambda x: torch.nn.init.constant(x, 1.), # everything else
}
init_all(model, init_funcs)
You can try with torch.nn.init.constant_(x, len(x.shape)) to check that they are appropriately initialized:
init_funcs = {
"default": lambda x: torch.nn.init.constant_(x, len(x.shape))
}
Here is the better way, just pass your whole model
import torch.nn as nn
def initialize_weights(model):
# Initializes weights according to the DCGAN paper
for m in model.modules():
if isinstance(m, (nn.Conv2d, nn.ConvTranspose2d, nn.BatchNorm2d)):
nn.init.normal_(m.weight.data, 0.0, 0.02)
# if you also want for linear layers ,add one more elif condition
Cuz I haven't had the enough reputation so far, I can't add a comment under
the answer posted by prosti in Jun 26 '19 at 13:16.
def reset_parameters(self):
init.kaiming_uniform_(self.weight, a=math.sqrt(3))
if self.bias is not None:
fan_in, _ = init._calculate_fan_in_and_fan_out(self.weight)
bound = 1 / math.sqrt(fan_in)
init.uniform_(self.bias, -bound, bound)
But I wanna point out that actually we know some assumptions in the paper of Kaiming He, Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification, are not appropriate, though it looks like the deliberately designed initialization method makes a hit in practice.
E.g., within the subsection of Backward Propagation Case, they assume that $w_l$ and $\delta y_l$ are independent of each other. But as we all known, take the score map $\delta y^L_i$ as an instance, it often is $y_i-softmax(y^L_i)=y_i-softmax(w^L_ix^L_i)$ if we use a typical cross entropy loss function objective.
So I think the true underlying reason why He's Initialization works well remains to unravel. Cuz everyone has witnessed its power on boosting deep learning training.
If you see a deprecation warning (@Fábio Perez)...
def init_weights(m):
if type(m) == nn.Linear:
torch.nn.init.xavier_uniform_(m.weight)
m.bias.data.fill_(0.01)
net = nn.Sequential(nn.Linear(2, 2), nn.Linear(2, 2))
net.apply(init_weights)
© 2022 - 2024 — McMap. All rights reserved.