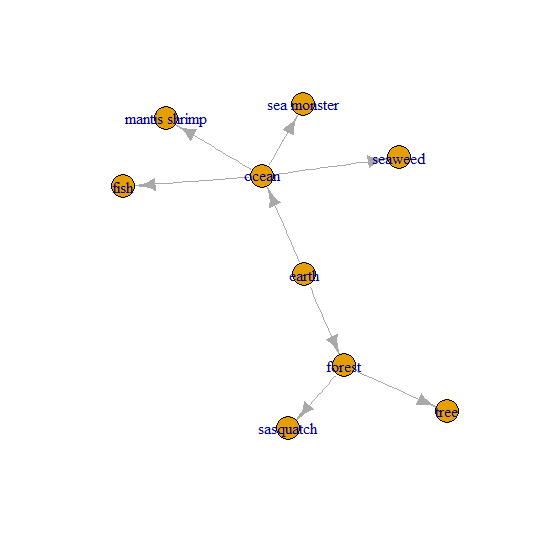
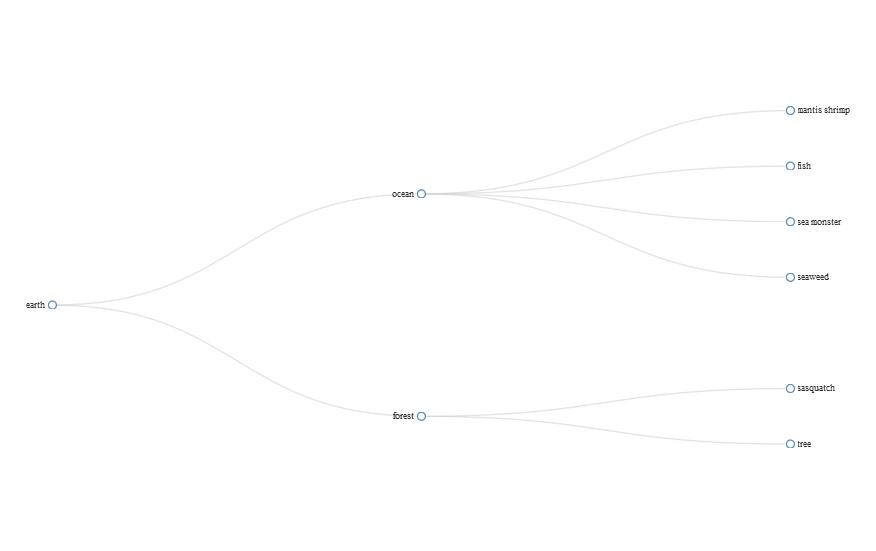
This is my first question; so please, be gentle.
I have some data that is in the form of :
library('networkD3')
Relationships<- data.frame(Parent=c("earth","earth","forest","forest","ocean","ocean","ocean","ocean"),
Child=c("ocean","forest","tree","sasquatch","fish","seaweed","mantis shrimp","sea monster"))
> Relationships
Parent Child
1 earth ocean
2 earth forest
3 forest tree
4 forest sasquatch
5 ocean fish
6 ocean seaweed
7 ocean mantis shrimp
8 ocean sea monster
Essentially this is a list of edges that can be used to make a network map:
net <- graph_from_data_frame(d = Relationships,
directed = T)
plot(net)
I would like to convert it to a form that could be used in the diagonalNetwork function, below.
Hierarchical_list <- list(name = "earth",
children = list(list(name = "ocean",
children = list(list(name = "mantis shrimp"),
list(name = "fish"),
list(name = "sea monster"),
list(name = "seaweed")
)),
list(name = "forest",
children = list(list(name = "sasquatch"),
list(name = "tree")
))
))
diagonalNetwork(Hierarchical_list)
Like this:
When I try to generate the list using this loop:
List_attempt <- list()
levels<- levels(factor(Relationships$Parent))
for(n in 1:length(levels)){
Children <- subset(Relationships, Relationships$Parent == levels[n], select = Child)
for(c in 1:length(Children)){
sublist <- as.list(Children)
List_attempt <- list(List_attempt, name = levels[n],children = sublist)
}
}
diagonalNetwork(List_attempt)
I get this error:
Error in FUN(X[[i]], ...) :
'options' must be a fully named list, or have no names (NULL)
1) Is there a better way of creating a list for diagonalNetwork?
2) Failing that; How can I modify my loops to kick out a list of the correct structure?
3) Is there a whole other function/package I should be using?
Thanks for any help you can give, I have been beating my head against this wall for a while. Feedback on better ways to ask questions on SO would also be welcomed.
Clarification:
A similar question is found here, Convert a data frame to a treeNetwork compatible list. However it relies on a data structure where the root is always in the first column, and its children are in subsequent columns, not a list of edges as in this question, which is commonly used in igraph.