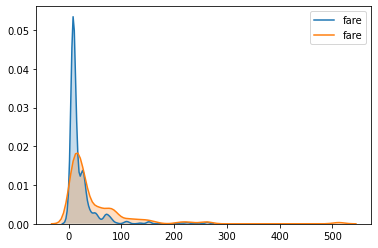
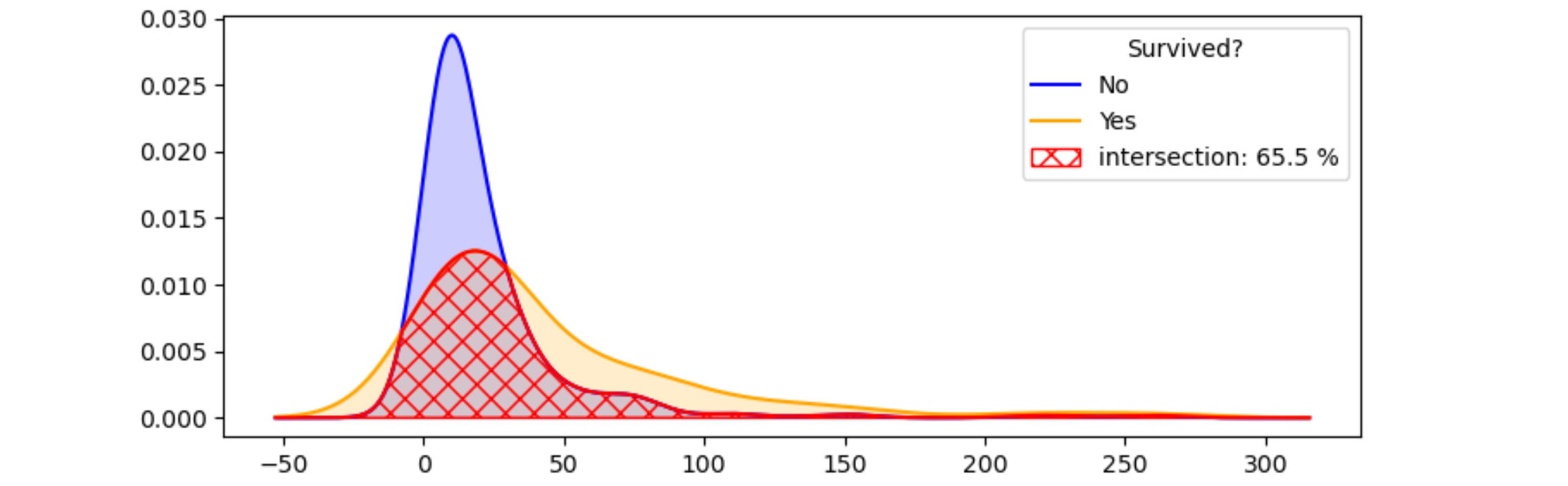
I was attempting to determine whether a feature is important or not base on its kde distribution for target variable. I am aware how to plot the kde plot and guess after looking at the plots, but is there a more formal doing this? Such as can we calculate the area of non overlapping area between two curves?
When I googled for the area between two curves there are many many links but none of them could solve my exact problem.
NOTE:
The main aim of this plot is to find whether the feature is important or not. So, please suggest me further if I am missing any hidden concepts here.
What I am trying to do is set some threshold such as 0.2, if the non-overlapping area > 0.2, then assert that the feature is important, otherwise not.
MWE:
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
df = sns.load_dataset('titanic')
x0 = df.loc[df['survived']==0,'fare']
x1 = df.loc[df['survived']==1,'fare']
sns.kdeplot(x0,shade=1)
sns.kdeplot(x1,shade=1)