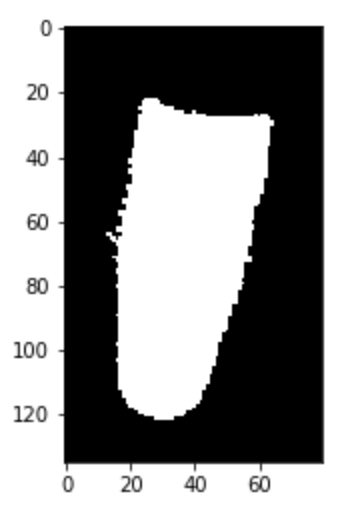
I assume, you want to keep the actual brain ("blob in the center") and get rid of the skull ("noise circular looking shape").
Unfortunately, you didn't show any code, so I'm not sure, what failed for you in using contours, but here's my suggestion:
import cv2
import numpy as np
# Read input
img = cv2.imread('images/t6igVVk.png', cv2.IMREAD_GRAYSCALE)
# Generate intermediate image; use morphological closing to keep parts of the brain together
inter = cv2.morphologyEx(img, cv2.MORPH_CLOSE, cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (5, 5)))
# Find largest contour in intermediate image
cnts, _ = cv2.findContours(inter, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_NONE)
cnt = max(cnts, key=cv2.contourArea)
# Output
out = np.zeros(img.shape, np.uint8)
cv2.drawContours(out, [cnt], -1, 255, cv2.FILLED)
out = cv2.bitwise_and(img, out)
cv2.imshow('img', img)
cv2.imshow('inter', inter)
cv2.imshow('out', out)
cv2.waitKey(0)
cv2.destroyAllWindows()
The final output out.png looks like this:
![Output]()
I do some morphological closing (morphologyEx, getStructuringElement) on the input image to keep the actual parts of the brain together. On that intermediate image, I look for the largest contour. In my findContours call, I use theRETR_EXTERNAL mode to only get all "external contours". That means, when later drawing this contour, it'll be also filled in the middle (i.e. the lateral ventricles). So, finally I just use a bitwise_and on both images to fix that. That also fixes the too large parts from the intermediate image as well.
Hope that helps!