I am using the ..count.. transformation in geom_bar and get the warning
position_stack requires non-overlapping x intervals when some of my categories have few counts.
This is best explained using some mock data (my data involves direction and windspeed and I retain names relating to that)
#make data
set.seed(12345)
FF=rweibull(100,1.7,1)*20 #mock speeds
FF[FF>60]=59
dir=sample.int(10,size=100,replace=TRUE) # mock directions
#group into speed classes
FFcut=cut(FF,breaks=seq(0,60,by=20),ordered_result=TRUE,right=FALSE,drop=FALSE)
# stuff into data frame & plot
df=data.frame(dir=dir,grp=FFcut)
ggplot(data=df,aes(x=dir,y=(..count..)/sum(..count..),fill=grp)) + geom_bar()
This works fine, and the resulting plot shows the frequency of directions grouped according to speed. It is of relevance that the velocity class with the fewest counts (here "[40,60)") will have 5 counts.
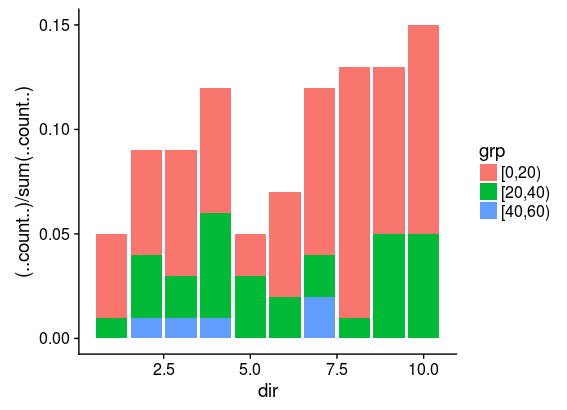
However more velocity classes leads to a warning. For instance, with
FFcut=cut(FF,breaks=seq(0,60,by=15),ordered_result=TRUE,right=FALSE,drop=FALSE)
the velocity class with the fewest counts (now "[45,60)") will have only 3 counts and ggplot2 will warn that
position_stack requires non-overlapping x intervals
and the plot will show data in this category spread out along the x axis.
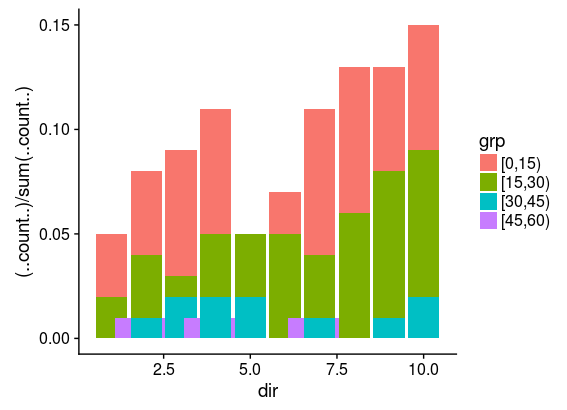 It seems that 5 is the minimum size for a group to have for this to work correctly.
It seems that 5 is the minimum size for a group to have for this to work correctly.
I would appreciate knowing if this is a feature or a bug in stat_bin (which geom_bar is using) or if I am simply abusing geom_bar.
Also, any suggestions how to get around this would be appreciated.
Sincerely

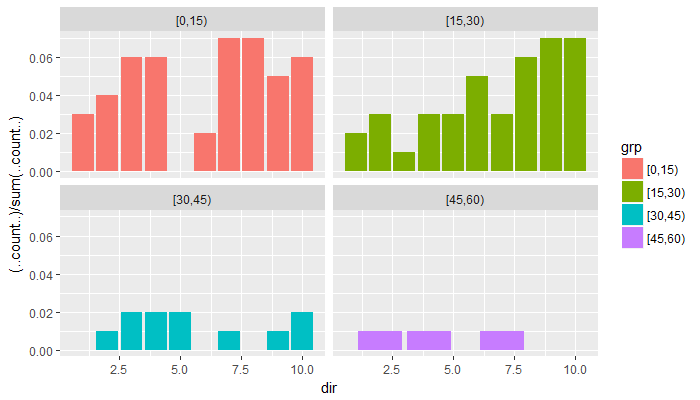
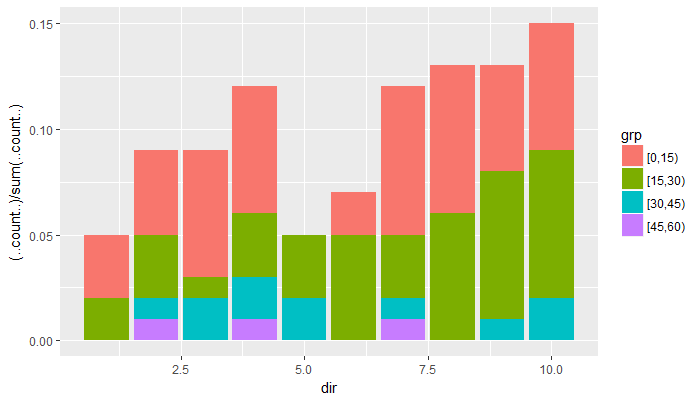

ggplot(data=df,aes(dir, fill=grp)) + geom_histogram(aes(y=(..count..)/sum(..count..)))– Transom