I am tying to bulk insert a dataframe to my postgres dB. Some columns in my dataframe are date types with NaT as a null value. Which is not supported by PostgreSQL, I've tried to replace NaT (using pandas) with other NULL type identifies but that did not work during my inserts.
I used df = df.where(pd.notnull(df), 'None') to replace all the NaTs, Example of errors that keep coming up due to datatype issues.
Error: invalid input syntax for type date: "None"
LINE 1: ...0,1.68757,'2022-11-30T00:29:59.679000'::timestamp,'None','20...
My driver and insert statement to postgresql dB:
def execute_values(conn, df, table):
"""
Using psycopg2.extras.execute_values() to insert the dataframe
"""
# Create a list of tupples from the dataframe values
tuples = [tuple(x) for x in df.to_numpy()]
# Comma-separated dataframe columns
cols = ','.join(list(df.columns))
# SQL quert to execute
query = "INSERT INTO %s(%s) VALUES %%s" % (table, cols)
cursor = conn.cursor()
try:
extras.execute_values(cursor, query, tuples)
conn.commit()
except (Exception, psycopg2.DatabaseError) as error:
print("Error: %s" % error)
conn.rollback()
cursor.close()
return 1
print("execute_values() done")
cursor.close()
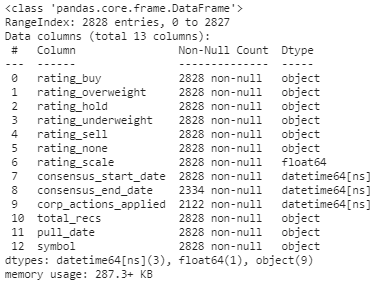
Info about my dataframe: for this case the culprits are the datetime columns only.
how is this commonly solved?