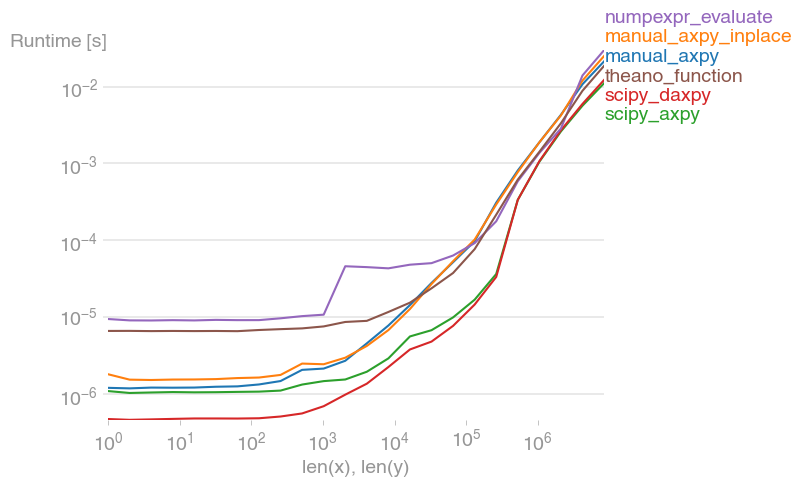
I compared different variants and found that you're not going wrong with SciPy's BLAS interface
scipy.linalg.blas.daxpy(x, y, len(x), a)
![enter image description here]()
Code to reproduce the plot:
import numexpr
import numpy as np
import perfplot
import scipy.linalg
import theano
a = 1.36
# theano preps
x = theano.tensor.vector()
y = theano.tensor.vector()
out = a * x + y
f = theano.function([x, y], out)
def setup(n):
x = np.random.rand(n)
y = np.random.rand(n)
return x, y
def manual_axpy(data):
x, y = data
return a * x + y
def manual_axpy_inplace(data):
x, y = data
out = a * x
out += y
return out
def scipy_axpy(data):
x, y = data
n = len(x)
axpy = scipy.linalg.blas.get_blas_funcs("axpy", arrays=(x, y))
axpy(x, y, n, a)
return y
def scipy_daxpy(data):
x, y = data
return scipy.linalg.blas.daxpy(x, y, len(x), a)
def numpexpr_evaluate(data):
x, y = data
return numexpr.evaluate("a * x + y")
def theano_function(data):
x, y = data
return f(x, y)
b = perfplot.bench(
setup=setup,
kernels=[
manual_axpy,
manual_axpy_inplace,
scipy_axpy,
scipy_daxpy,
numpexpr_evaluate,
theano_function,
],
n_range=[2 ** k for k in range(24)],
equality_check=None,
xlabel="len(x), len(y)",
)
# b.save("out.png")
b.show()

numpy.add(... out = C)? No extra array created that way. – Remingtoncommentsand question being tagged asmemorysuggests itsmemorythat you are focussing on. If that's the case,numpy.adduses no extra memory, as mentioned earlier as well. – Remingtonnumpy'suse of memory. If it's speed you are worried about, try several options on realistic arrays and see it they make any difference. – Stockbroker