I'm looking for a way to remove isolated white pixels from a binary image using OpenCV. A similar question (OpenCV get rid of isolated pixels) has a bunch of "answers" but none seem to work for me. I've tried various combinations of opening and closing without success as well.
The article here:
https://homepages.inf.ed.ac.uk/rbf/HIPR2/hitmiss.htm
Suggests I can use the hit-or-miss operation for exactly this purpose:
1 is used to locate isolated points in a binary image
And that the reason why is that 0s are interpreted differently than when they are used with erosion/dilation directly (where 0s are interpreted as "don't care's" rather than "not white" which is basically what I'm after). However, using this kernel simply renders the original image.
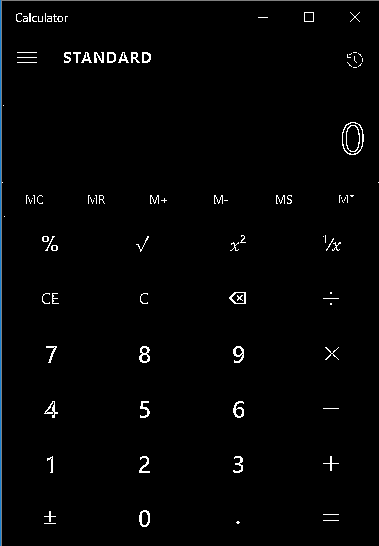
My input image is this:
You'll notice there's a few white pixels near the left-hand side of the image which I'd like to get rid of.
Here's the code:
kernel = np.array([ [0, 0, 0],
[0, 1, 0],
[0, 0, 0]],np.uint8)
hitormiss = cv2.morphologyEx(input_image, cv2.MORPH_HITMISS, kernel)
cv2.imshow('hitormiss', hitormiss)
What is the right way of removing isolated pixels like these?
Update: Alexander's answer works like a charm and is the fastest solution. The other answer provides a solution too, which is to use the cv2.connectedComponents function, but it is much more processor-intensive. Here's a function that uses this approach:
def remove_isolated_pixels(self, image):
connectivity = 8
output = cv2.connectedComponentsWithStats(image, connectivity, cv2.CV_32S)
num_stats = output[0]
labels = output[1]
stats = output[2]
new_image = image.copy()
for label in range(num_stats):
if stats[label,cv2.CC_STAT_AREA] == 1:
new_image[labels == label] = 0
return new_image

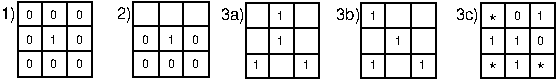

0,0,0 0,1,0 0,0,0kernel and it immediately isolated your noisy pixels. I notice your image is palettised - have you checked the actual values in your input image where you expect to find the noise - I mean printed out their actual values and the 8 surrounding pixels? – Initial