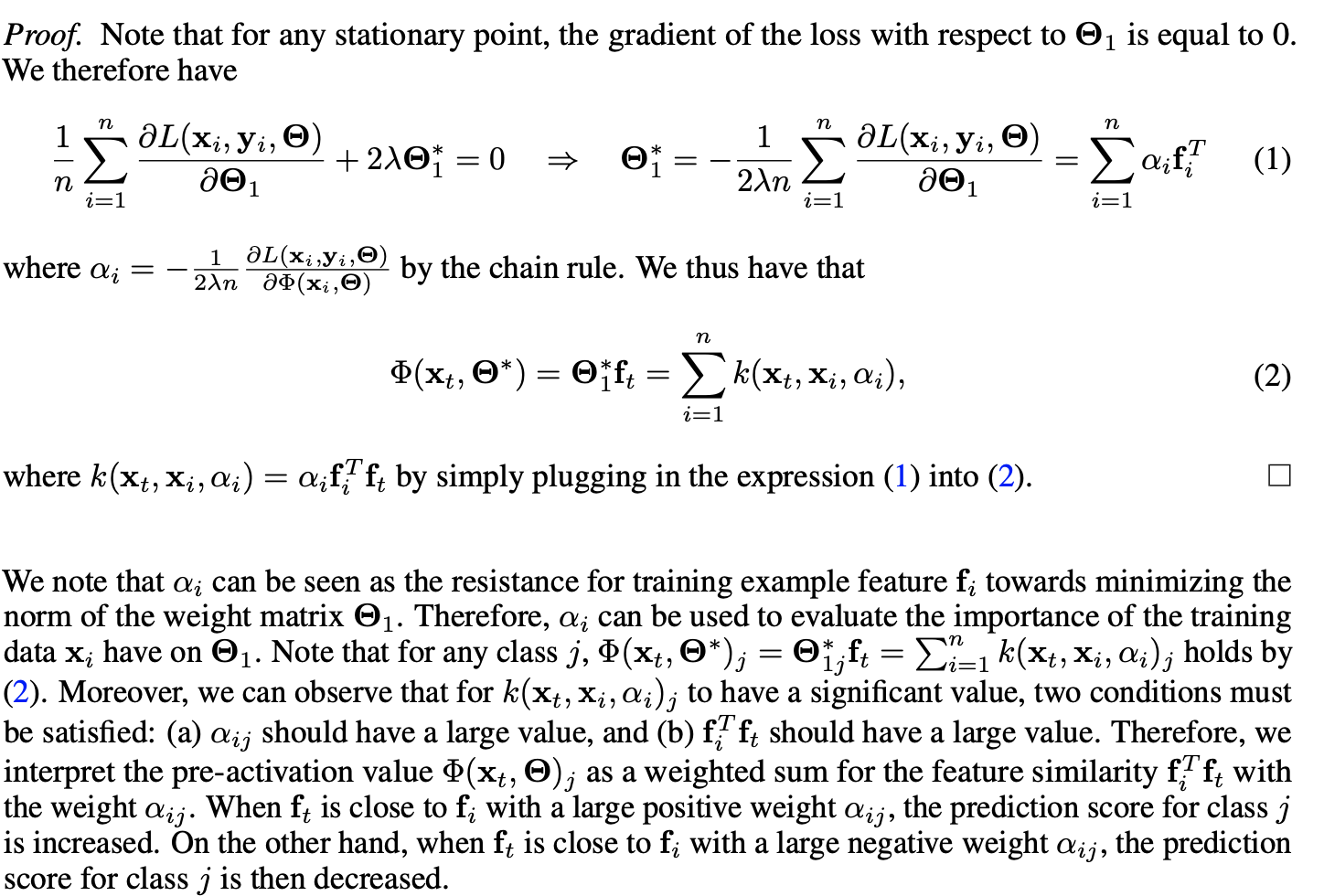
I am using tf.GradientTape().gradient() to compute a representer point, which can be used to compute the "influence" of a given training example on a given test example. A representer point for a given test example x_t and training example x_i is computed as the dot product of their feature representations, f_t and f_i, multiplied by a weight alpha_i.
Note: The details of this approach are not necessary for understanding the question, since the main issue is getting gradient tape to work. That being said, I have included a screenshot of the some of the details below for anyone who is interested.
Computing alpha_i requires differentiation, since it is expressed as the following:
In the equation above L is the standard loss function (categorical cross-entropy for multiclass classification) and phi is the pre-softmax activation output (so its length is the number of classes). Furthermore alpha_i can be further broken up into alpha_ij, which is computed with respect to a specific class j. Therefore, we just obtain the pre-softmax output phi_j corresponding to the predicted class of the test example (class with highest final prediction).
I have created a simple setup with MNIST and have implemented the following:
def simple_mnist_cnn(input_shape = (28,28,1)):
input = Input(shape=input_shape)
x = layers.Conv2D(32, kernel_size=(3, 3), activation="relu")(input)
x = layers.MaxPooling2D(pool_size=(2, 2))(x)
x = layers.Conv2D(64, kernel_size=(3, 3), activation="relu")(x)
x = layers.MaxPooling2D(pool_size=(2, 2))(x)
x = layers.Flatten()(x) # feature representation
output = layers.Dense(num_classes, activation=None)(x) # presoftmax activation output
activation = layers.Activation(activation='softmax')(output) # final output with activation
model = tf.keras.Model(input, [x, output, activation], name="mnist_model")
return model
Now assume the model is trained, and I want to compute the influence of a given train example on a given test example's prediction, perhaps for model understanding/debugging purposes.
with tf.GradientTape() as t1:
f_t, _, pred_t = model(x_t) # get features for misclassified example
f_i, presoftmax_i, pred_i = model(x_i)
# compute dot product of feature representations for x_t and x_i
dotps = tf.reduce_sum(
tf.multiply(f_t, f_i))
# get presoftmax output corresponding to highest predicted class of x_t
phi_ij = presoftmax_i[:,np.argmax(pred_t)]
# y_i is actual label for x_i
cl_loss_i = tf.keras.losses.categorical_crossentropy(pred_i, y_i)
alpha_ij = t1.gradient(cl_loss_i, phi_ij)
# note: alpha_ij returns None currently
k_ij = tf.reduce_sum(tf.multiply(alpha_i, dotps))
The code above gives the following error, since alpha_ij is None: ValueError: Attempt to convert a value (None) with an unsupported type (<class 'NoneType'>) to a Tensor.. However, if I change t1.gradient(cl_loss_i, phi_ij) -> t1.gradient(cl_loss_i, presoftmax_i), it no longer returns None. Not sure why this is the case? Is there an issue with computing gradients on sliced tensors? Is there an issue with "watching" too many variables? I haven't worked much with gradient tape so I'm not sure what the fix is, but would appreciate help.

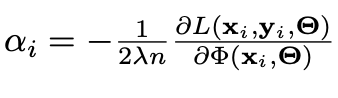
np.argmax(pred_t)to a fixed index (e.g. 0), and the issue still persisted. – Danie