The short answer is, if you're interested in coefficients of a polynomial in standard power basis, you'd be better off using CubicSpline (and see this discussion):
cu = scipy.interpolate.CubicSpline(x_sample, d_sample)
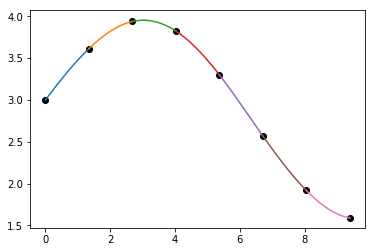
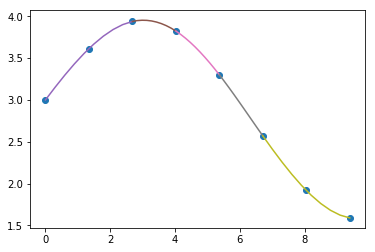
plt.plot(x_sample, y_sample, 'ko')
for i in range(len(cu.x)-1):
xs = np.linspace(cu.x[i], cu.x[i+1], 100)
plt.plot(xs, np.polyval(cu.c[:,i], xs - cu.x[i]))
![cubic spline piecewise]()
And to answer your question, you could instead create a piecewise function from here using numpy.piecewise, the breakpoints in cu.x and the coefficients in cu.c, and either directly code the polynomial expressions yourself or use numpy.polyval. For example,
cu.c[:,0] # coeffs for 0th segment
# array([-0.01316353, -0.02680068, 0.51629024, 3. ])
# equivalent ways to code polynomial for this segment
f0 = lambda x: cu.c[0,0]*(x-x[0])**3 + cu.c[1,0]*(x-x[0])**2 + cu.c[2,0]*(x-x[0]) + cu.c[3,0]
f0 = lambda x: [cu.c[i,0]*(x-x[0])**(3-i) for i in range(4)]
# ... or getting values directly from x's
y0 = np.polyval(cu.c[:,0], xs - cu.x[0])
LONGER ANSWER:
There are a few points of potential confusion here:
UnivariateSpline fits a B-spline basis, so the coefficients are not the same as a standard polynomial power basis- In order to convert from B-spline, we can use
PPoly.from_spline, but unfortunately UnivariateSpline returns a truncated list of knots and coefficients that won't play with this function. We can resolve this problem by accessing the internal data of the spline object, which is a little taboo.
- Also, the coefficient matrix
c (whether from UnivariateSpline or CubicSpline) is in reverse degree order and assumes you are "centering" yourself, e.g. the coefficient at c[k,i] belongs to c[k,i]*(x-x[i])^(3-k).
Given your setup, note that if instead of using the UnivariateSpline wrapper, we directly fit with splrep and no smoothing (s=0), we can grab the tck (knots-coefficients-degree) tuple and send it to the PPoly.from_spline function and get the coefficients we want:
tck = scipy.interpolate.splrep(x_sample, d_sample, s=0)
tck
# (array([0. , 0. , 0. , 0. , 2.68456376,
# 4.02684564, 5.36912752, 6.7114094 , 9.39597315, 9.39597315,
# 9.39597315, 9.39597315]),
# array([3. , 3.46200469, 4.05843704, 3.89649312, 3.33792889,
# 2.29435138, 1.65015175, 1.59021688, 0. , 0. ,
# 0. , 0. ]),
# 3)
p = scipy.interpolate.PPoly.from_spline(tck)
p.x.shape # breakpoints in unexpected shape
# (12,)
p.c.shape # polynomial coeffs in unexpected shape
# (4, 11)
Notice the weird repeated breakpoints in tck and again in p.x: this is a FITPACK thing (the algorithm running all this).
If we try to send a tck tuple from UnivariateSpline with (s.get_knots(), s.get_coeffs(), 3), we are missing those repeats, so from_spline doesn't work. Checking out the source though it appears the full vector is stored in self._data, so we can do
s = scipy.interpolate.UnivariateSpline(x_sample, d_sample, s=0)
tck = (s._data[8], s._data[9], 3)
p = scipy.interpolate.PPoly.from_spline(tck)
and get the same as before. To check these coefficients work:
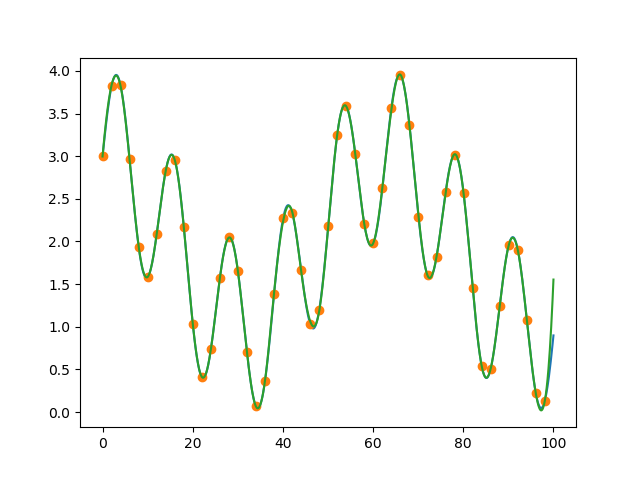
plt.plot(x_sample, d_sample, 'o')
for i in range(len(p.x)-1):
xs = np.linspace(p.x[i], p.x[i+1], 10)
plt.plot(xs, np.polyval(p.c[:,i], xs - p.x[i]))
![piecewise spline]()
Note numpy.polyval wants reverse order for coeffs so we can pass p.c as-is.

PPoly.from_spline. Once you have the coeffs, it's a simple application of, for example,np.polyevalandnp.piecewise. Have a look at this discussion on the trickiness of conversion. – UpwardPPoly.from_splinepart? I do not have to useUnivariateSpline, but any spline is fine that gives mefiI could then indeed feed intopiecewise. – Barytone