I created a graph using geom_line and geom_point via ggplot. I want my axes to meet at (0,0) and I want my lines and data points to be in front of the axes instead of behind as shown:

I've tried:
- coord_cartesian(clip = 'off')
- putting geom_line and geom_point at the end
- creating a base graph then add geom_line and geom_point
- playing around with the functions of coord_cartesian
- manually setting xlim =c(-0.1, 25) and ylim=c(-0.1, 1500)
data7 is as follows:
Treatment Days N mean sd se
1 1 0 7 204.7000000 41.579963 15.7157488
2 1 2 7 255.0571429 41.116617 15.5406205
3 1 5 7 290.6000000 49.506498 18.7116974
4 1 8 7 330.8142857 49.044144 18.5369442
5 1 12 7 407.5142857 95.584194 36.1274294
6 1 15 7 540.8571429 164.299390 62.0993323
7 1 19 7 737.5285714 308.786359 116.7102736
8 1 21 7 978.4571429 502.506726 189.9296898
9 2 0 7 205.7428571 46.902482 17.7274721
10 2 2 7 227.5571429 47.099889 17.8020846
11 2 5 7 232.4857143 59.642922 22.5429054
12 2 8 7 247.9857143 66.478529 25.1265220
13 2 12 7 272.0428571 79.173162 29.9246423
14 2 15 7 289.1142857 82.847016 31.3132288
15 2 19 7 312.3857143 105.648591 39.9314140
16 2 21 7 334.7142857 121.569341 45.9488920
17 3 0 7 212.2285714 47.549263 17.9719320
18 3 2 7 235.4142857 52.689671 19.9148237
19 3 5 7 177.0714286 54.895225 20.7484447
20 3 8 7 205.2571429 72.611451 27.4445489
21 3 12 7 247.8142857 119.369558 45.1174522
22 3 15 7 280.4285714 140.825847 53.2271669
23 3 19 7 366.9142857 210.573799 79.5894149
24 3 21 7 451.0428571 289.240793 109.3227438
25 4 0 7 211.6857143 24.329161 9.1955587
26 4 2 7 227.8428571 28.762525 10.8712127
27 4 5 7 205.9428571 49.148919 18.5765451
28 4 8 7 153.1142857 25.189246 9.5206399
29 4 12 7 128.2571429 43.145910 16.3076210
30 4 15 7 104.1714286 45.161662 17.0695038
31 4 19 7 85.4714286 51.169708 19.3403318
32 4 21 7 66.9000000 52.724567 19.9280133
33 5 0 7 216.7857143 39.957829 15.1026398
34 5 2 7 212.2000000 27.037135 10.2190765
35 5 5 7 115.5000000 37.094070 14.0202405
36 5 8 7 46.1000000 34.925492 13.2005952
37 5 12 7 29.3142857 24.761222 9.3588621
38 5 15 6 10.0666667 13.441974 5.4876629
39 5 19 6 6.4000000 11.692733 4.7735382
40 5 21 6 5.3666667 12.662017 5.1692467
41 6 0 7 206.6857143 40.359155 15.2543269
42 6 2 7 197.0428571 40.608327 15.3485048
43 6 5 7 106.2142857 58.279654 22.0276388
44 6 8 7 46.0571429 62.373014 23.5747833
45 6 12 7 31.7571429 49.977457 18.8897031
46 6 15 7 28.1142857 45.437995 17.1739480
47 6 19 7 26.2857143 38.414946 14.5194849
48 6 21 7 32.7428571 53.203003 20.1088450
49 7 0 7 193.2000000 37.300447 14.0982437
50 7 2 7 133.2428571 26.462606 10.0019250
51 7 5 7 3.8142857 7.445900 2.8142857
52 7 8 7 0.7142857 1.496026 0.5654449
53 7 12 7 0.0000000 0.000000 0.0000000
54 7 15 7 0.0000000 0.000000 0.0000000
55 7 19 7 0.0000000 0.000000 0.0000000
56 7 21 7 0.0000000 0.000000 0.0000000
My code is as follows:
ggplot(data7, aes(Days, mean, color=Treatment)) +
geom_line() +
geom_errorbar(aes(ymin=mean-se, ymax=mean+se), width=0.5, size= 0.25) +
geom_point(size=2.5) +
scale_colour_hue(limits = c("1", "2", "3", "4", "5", "6", "7")) +
scale_x_continuous(expand = c(0, 0), limits = c(0, NA), breaks = scales::pretty_breaks(n = 10)) +
scale_y_continuous(expand = c(0, 0), limits = c(0, NA), breaks = scales::pretty_breaks(n = 8)) +
theme_classic() +
theme(axis.text = element_text(color = "#000000"), plot.title = element_text(hjust = 0.5)) +
coord_cartesian(clip = 'off')

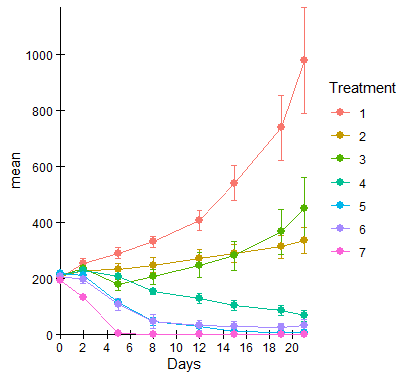
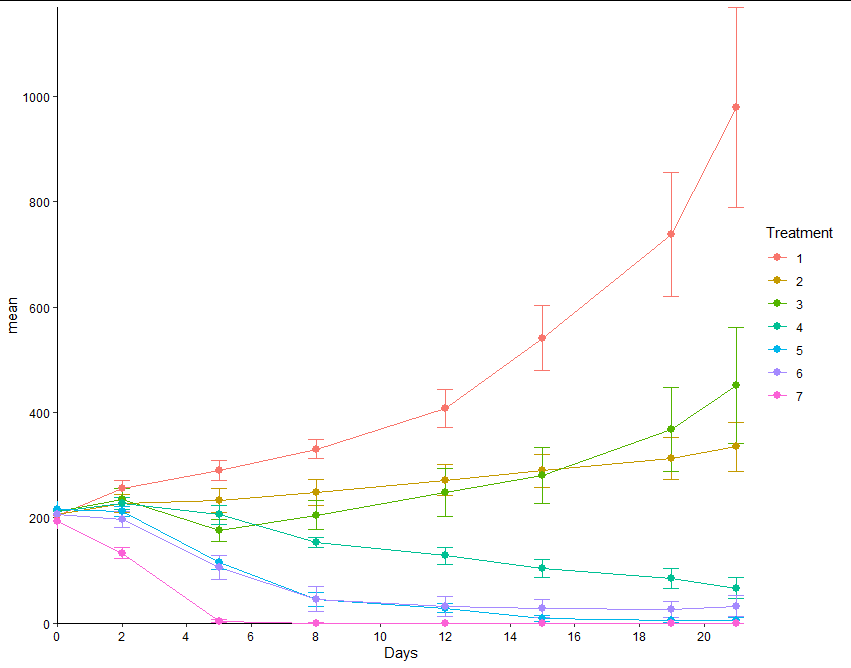
x=0, it's a little left of it. – Lantz0,0intersection :-), hope it works (or at least helps). – Lantz