Summary (tldr)
If you want the 'python3' kernel to always run the Python installation from the environment where it is launched, delete the User 'python3' kernel, which is taking precedence over whatever the current environment is with:
jupyter kernelspec remove python3
Full Solution
I am going to post an alternative and simpler solution for the following case:
- You have created a conda environment
- This environment has jupyter installed (which also installs ipykernel)
- When you run the command
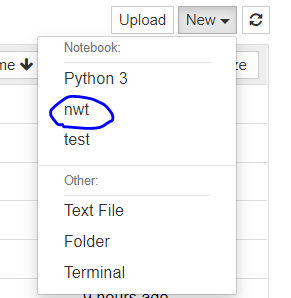
jupyter notebook and create a new notebook by clicking 'python3' in the 'New' dropdown menu, that notebook executes python from the base environment and not from the current environment.
- You would like it so that launching a new notebook with 'python3' within any environment executes the Python version from that environment and NOT the base
I am going to use the name 'test_env' for the environment for the rest of the solution. Also, note that 'python3' is the name of the kernel.
The currently top-voted answer does work, but there is an alternative. It says to do the following:
python -m ipykernel install --user --name test_env --display-name "Python (test_env)"
This will give you the option of using the test_env environment regardless of what environment you launch jupyter notebook from. But, launching a notebook with 'python3' will still use the Python installation from the base environment.
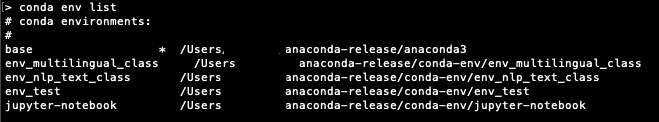
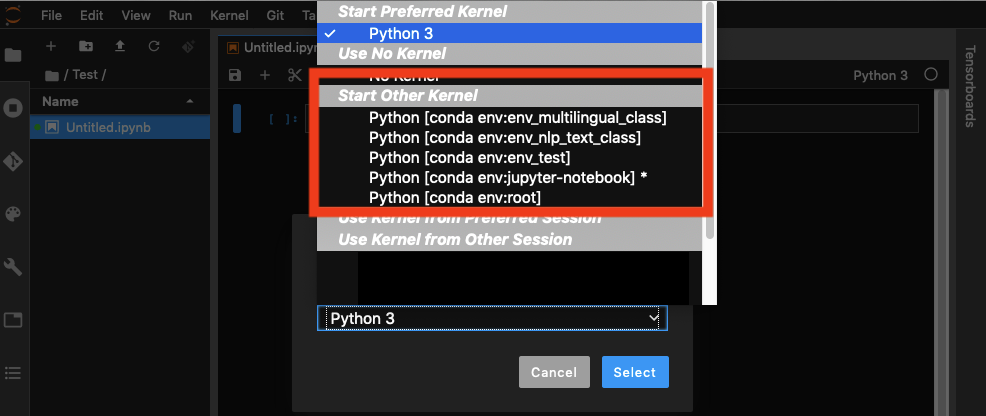
What likely is happening is that there is a user python3 kernel that exists. Run the command jupyter kernelspec list to list all of your environments. For instance, if you have a mac you will be returned the following (my user name is Ted).
python3 /Users/Ted/Library/Jupyter/kernels/python3
What Jupyter is doing here is searching through three different paths looking for kernels. It goes from User, to Env, to System. See this document for more details on the paths it searches for each operating system.
The two kernels above are both in the User path, meaning they will be available regardless of the environment that you launch a jupyter notebook from. This also means that if there is another 'python3' kernel at the environment level, then you will never be able to access it.
To me, it makes more sense that choosing the 'python3' kernel from the environment you launched the notebook from should execute Python from that environment.
You can check to see if you have another 'python3' environment by looking in the Env search path for your OS (see the link to the docs above). For me (on my mac), I issued the following command:
ls /Users/Ted/anaconda3/envs/test_env/share/jupyter/kernels
And I indeed had a 'python3' kernel listed there.
Thanks to this GitHub issue comment (look at the first response), you can remove the User 'python3' environment with the following command:
jupyter kernelspec remove python3
Now when you run jupyter kernelspec list, assuming the test_env is still active, you will get the following:
python3 /Users/Ted/anaconda3/envs/test_env/share/jupyter/kernels/python3
Notice that this path is located within the test_env directory. If you create a new environment, install jupyter, activate it, and list the kernels, you will get another 'python3' kernel located in its environment path.
The User 'python3' kernel was taking precedence over any of the Env 'python3' kernels. By removing it, the active environment 'python3' kernel was exposed and able to be chosen every time. This eliminates the need to manually create kernels. It also makes more sense in terms of software development where one would want to isolate themselves into a single environment. Running a kernel that is different from the host environment doesn't seem natural.
It also seems that this User 'python3' is not installed for everyone by default, so not everyone is confronted by this issue.


conda install ipykernelseems to installjupyterin the environment... Am I missing something? – Exuviatenb_condais used or if the kernel is setup manually as suggested in the question. Otherwise it will actually mess up things quite a lot. The executablejupyterwill point to an executable inside the environment, but system'sjupyter-notebookwill be started (if installed) and therefore not use the environment with the default kernel. – Tobolsk