If speed is what you need and extra dependencies are not a problem, you maybe find numba quite useful (now it is pretty easy to install, on any platform). The classic ray_tracing approach you proposed can be easily ported to numba by using numba @jit decorator and casting the polygon to a numpy array. The code should look like:
@jit(nopython=True)
def ray_tracing(x,y,poly):
n = len(poly)
inside = False
p2x = 0.0
p2y = 0.0
xints = 0.0
p1x,p1y = poly[0]
for i in range(n+1):
p2x,p2y = poly[i % n]
if y > min(p1y,p2y):
if y <= max(p1y,p2y):
if x <= max(p1x,p2x):
if p1y != p2y:
xints = (y-p1y)*(p2x-p1x)/(p2y-p1y)+p1x
if p1x == p2x or x <= xints:
inside = not inside
p1x,p1y = p2x,p2y
return inside
The first execution will take a little longer than any subsequent call:
%%time
polygon=np.array(polygon)
inside1 = [numba_ray_tracing_method(point[0], point[1], polygon) for
point in points]
CPU times: user 129 ms, sys: 4.08 ms, total: 133 ms
Wall time: 132 ms
Which, after compilation will decrease to:
CPU times: user 18.7 ms, sys: 320 µs, total: 19.1 ms
Wall time: 18.4 ms
If you need speed at the first call of the function you can then pre-compile the code in a module using pycc. Store the function in a src.py like:
from numba import jit
from numba.pycc import CC
cc = CC('nbspatial')
@cc.export('ray_tracing', 'b1(f8, f8, f8[:,:])')
@jit(nopython=True)
def ray_tracing(x,y,poly):
n = len(poly)
inside = False
p2x = 0.0
p2y = 0.0
xints = 0.0
p1x,p1y = poly[0]
for i in range(n+1):
p2x,p2y = poly[i % n]
if y > min(p1y,p2y):
if y <= max(p1y,p2y):
if x <= max(p1x,p2x):
if p1y != p2y:
xints = (y-p1y)*(p2x-p1x)/(p2y-p1y)+p1x
if p1x == p2x or x <= xints:
inside = not inside
p1x,p1y = p2x,p2y
return inside
if __name__ == "__main__":
cc.compile()
Build it with python src.py and run:
import nbspatial
import numpy as np
lenpoly = 100
polygon = [[np.sin(x)+0.5,np.cos(x)+0.5] for x in
np.linspace(0,2*np.pi,lenpoly)[:-1]]
# random points set of points to test
N = 10000
# making a list instead of a generator to help debug
points = zip(np.random.random(N),np.random.random(N))
polygon = np.array(polygon)
%%time
result = [nbspatial.ray_tracing(point[0], point[1], polygon) for point in points]
CPU times: user 20.7 ms, sys: 64 µs, total: 20.8 ms
Wall time: 19.9 ms
In the numba code I used:
'b1(f8, f8, f8[:,:])'
In order to compile with nopython=True, each var needs to be declared before the for loop.
In the prebuild src code the line:
@cc.export('ray_tracing' , 'b1(f8, f8, f8[:,:])')
Is used to declare the function name and its I/O var types, a boolean output b1 and two floats f8 and a two-dimensional array of floats f8[:,:] as input.
Edit Jan/4/2021
For my use case, I need to check if multiple points are inside a single polygon - In such a context, it is useful to take advantage of numba parallel capabilities to loop over a series of points. The example above can be changed to:
from numba import jit, njit
import numba
import numpy as np
@jit(nopython=True)
def pointinpolygon(x,y,poly):
n = len(poly)
inside = False
p2x = 0.0
p2y = 0.0
xints = 0.0
p1x,p1y = poly[0]
for i in numba.prange(n+1):
p2x,p2y = poly[i % n]
if y > min(p1y,p2y):
if y <= max(p1y,p2y):
if x <= max(p1x,p2x):
if p1y != p2y:
xints = (y-p1y)*(p2x-p1x)/(p2y-p1y)+p1x
if p1x == p2x or x <= xints:
inside = not inside
p1x,p1y = p2x,p2y
return inside
@njit(parallel=True)
def parallelpointinpolygon(points, polygon):
D = np.empty(len(points), dtype=numba.boolean)
for i in numba.prange(0, len(D)):
D[i] = pointinpolygon(points[i,0], points[i,1], polygon)
return D
Note: pre-compiling the above code will not enable the parallel capabilities of numba (parallel CPU target is not supported by pycc/AOT compilation) see: https://github.com/numba/numba/issues/3336
Test:
import numpy as np
lenpoly = 100
polygon = [[np.sin(x)+0.5,np.cos(x)+0.5] for x in np.linspace(0,2*np.pi,lenpoly)[:-1]]
polygon = np.array(polygon)
N = 10000
points = np.random.uniform(-1.5, 1.5, size=(N, 2))
For N=10000 on a 72 core machine, returns:
%%timeit
parallelpointinpolygon(points, polygon)
# 480 µs ± 8.19 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
Edit 17 Feb '21:
- fixing loop to start from
0 instead of 1 (thanks @mehdi):
for i in numba.prange(0, len(D))
Edit 20 Feb '21:
Follow-up on the comparison made by @mehdi, I am adding a GPU-based method below. It uses the point_in_polygon method, from the cuspatial library:
import numpy as np
import cudf
import cuspatial
N = 100000002
lenpoly = 1000
polygon = [[np.sin(x)+0.5,np.cos(x)+0.5] for x in
np.linspace(0,2*np.pi,lenpoly)]
polygon = np.array(polygon)
points = np.random.uniform(-1.5, 1.5, size=(N, 2))
x_pnt = points[:,0]
y_pnt = points[:,1]
x_poly =polygon[:,0]
y_poly = polygon[:,1]
result = cuspatial.point_in_polygon(
x_pnt,
y_pnt,
cudf.Series([0], index=['geom']),
cudf.Series([0], name='r_pos', dtype='int32'),
x_poly,
y_poly,
)
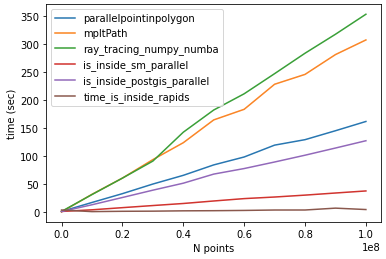
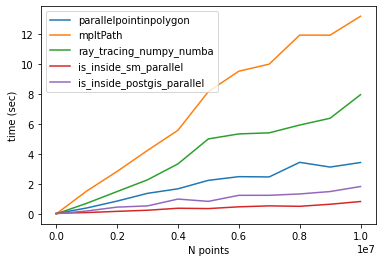
Following @Mehdi comparison. For N=100000002 and lenpoly=1000 - I got the following results:
time_parallelpointinpolygon: 161.54760098457336
time_mpltPath: 307.1664695739746
time_ray_tracing_numpy_numba: 353.07356882095337
time_is_inside_sm_parallel: 37.45389246940613
time_is_inside_postgis_parallel: 127.13793849945068
time_is_inside_rapids: 4.246025562286377
![point in poligon methods comparison, #poins: 10e07]()
hardware specs:
- CPU Intel xeon E1240
- GPU Nvidia GTX 1070
Notes:
The cuspatial.point_in_poligon method, is quite robust and powerful, it offers the ability to work with multiple and complex polygons (I guess at the expense of performance)
The numba methods can also be 'ported' on the GPU - it will be interesting to see a comparison which includes a porting to cuda of fastest method mentioned by @Mehdi (is_inside_sm).

polygon = np.array([[0, 0],[1, 0],[ 0, 1],[ 1, 1]])points = np.array([[0.5, 0.5]])Only matplotlib.path returns the correct result. – Vachon