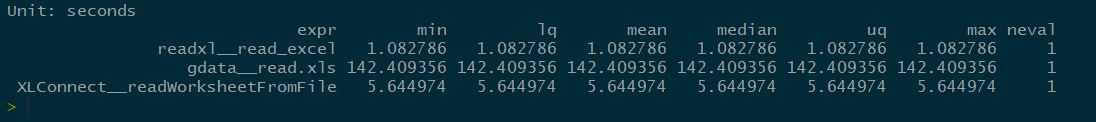
I have multiple .xls (~100MB) files from which I would like to load multiple sheets (from each) into R as a dataframe. I have tried various functions, such as xlsx::xlsx2 and XLConnect::readWorksheetFromFile, both of which always run for a very long time (>15 mins) and never finish and I have to force-quit RStudio to keep working.
I also tried gdata::read.xls, which does finish, but it takes more than 3 minutes per one sheet and it cannot extract multiple sheets at once (which would be very helpful to speed up my pipeline) like XLConnect::loadWorkbook can.
The time it takes these functions to execute (and I am not even sure the first two would ever finish if I let them go longer) is way too long for my pipeline, where I need to work with many files at once. Is there a way to get these to go/finish faster?
In several places, I have seen a recommendation to use the function readxl::read_xls, which seems to be widely recommended for this task and should be faster per sheet. This one, however, gives me an error:
> # Minimal reproducible example:
> setwd("/Users/USER/Desktop")
> library(readxl)
> data <- read_xls(path="test_file.xls")
Error:
filepath: /Users/USER/Desktop/test_file.xls
libxls error: Unable to open file
I also did some elementary testing to make sure the file exists and is in the correct format:
> # Testing existence & format of the file
> file.exists("test_file.xls")
[1] TRUE
> format_from_ext("test_file.xls")
[1] "xls"
> format_from_signature("test_file.xls")
[1] "xls"
The test_file.xls used above is available here.
Any advice would be appreciated in terms of making the first functions run faster or the read_xls run at all - thank you!
UPDATE (12/14/2019):
It seems that some users are able to open the file above using the readxl::read_xls function, while others are not, both on Mac and Windows, using the most up to date versions of R, Rstudio, and readxl. The issue has been posted on the readxl GitHub and has not been resolved yet.
UPDATE (4/14/2023):
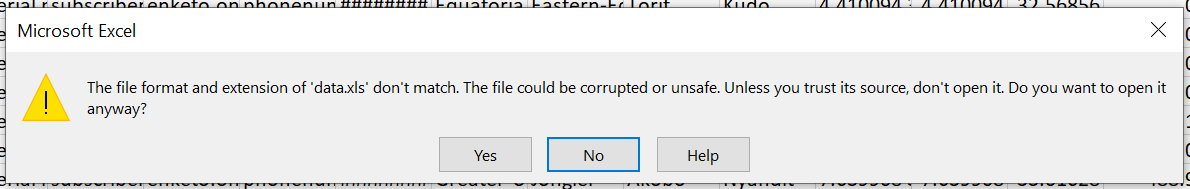
The above GitHub issue became a combination of various problems and yielded one possible solution, which was opening and closing the file in MS Excel first, which subsequently made it readable by readxl::read_xls. Understandably, though, this is not the best solution. The above issue was closed without a better resolution and instead, a related issue was opened in the libxls GitHub, so far without a resolution.


libxls error: Unable to open file– Providential