Important to note that the solution code in another answer here is incorrect. It does not only fail to get the linear time by using the median of medians algorithm which is strictly required for such time complexity, but it uses average instead of median and divides the list into potentially unequal parts which could cause an infinite loop or O(mn) time and both of which violate the assumptions of Camerini's original algorithm. Very limited test cases are not adequate to prove such an algorithm works, it takes a good amount of them before empirical verification can be considered adequate.
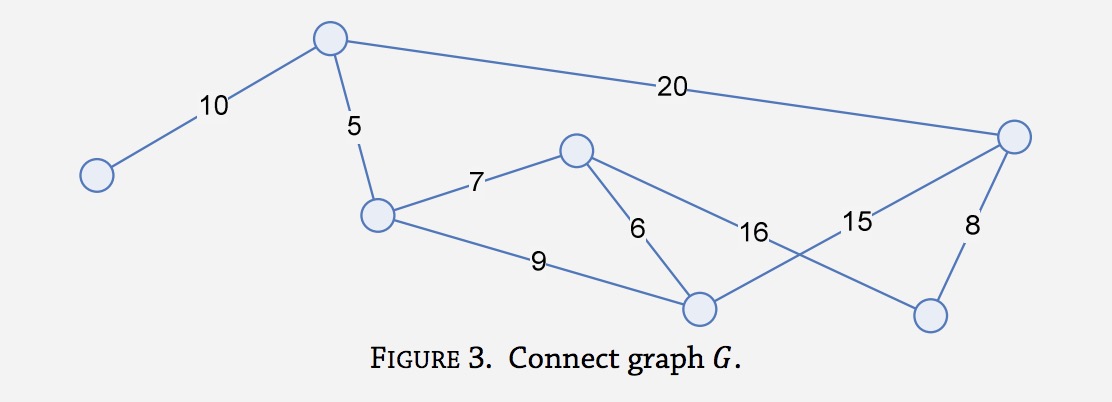
Here is Python code which will solve the problem using Camerini's algorithm with proper time complexity. The median of medians algorithm is quite long but the only way to achieve it. The Wikipedia example as well as a visualization function using graphviz is provided. It is assumed you are talking about undirected graphs per the question referencing Kruskal with O(m log n). Some unnecessary adjacency matrix to edge list conversions which is O(n^2) is here but it can be easily optimized out.
def adj_mat_to_edge_list(adj_mat):
#undirected graph needs reflexivity check
return {i+1:[j+1 for j, y in enumerate(row) if not y is None or not adj_mat[j][i] is None]
for i, row in enumerate(adj_mat)}
def adj_mat_to_costs(adj_mat):
return {(i+1, j+1): adj_mat[i][j] for i, row in enumerate(adj_mat)
for j, y in enumerate(row) if not y is None or not adj_mat[j][i] is None}
def partition5(l, left, right):
i = left + 1
while i <= right:
j = i
while j > left and l[j-1] > l[j]:
l[j], l[j-1] = l[j-1], l[j]
j -= 1
i += 1
return (left + right) // 2
def pivot(l, left, right):
if right - left < 5: return partition5(l, left, right)
for i in range(left, right+1, 5):
subRight = i + 4
if subRight > right: subRight = right
median5 = partition5(l, i, subRight)
l[median5], l[left + (i-left) // 5] = l[left + (i-left) // 5], l[median5]
mid = (right - left) // 10 + left + 1
return medianofmedians(l, left, left + (right - left) // 5, mid)
def partition(l, left, right, pivotIndex, n):
pivotValue = l[pivotIndex]
l[pivotIndex], l[right] = l[right], l[pivotIndex]
storeIndex = left
for i in range(left, right):
if l[i] < pivotValue:
l[storeIndex], l[i] = l[i], l[storeIndex]
storeIndex += 1
storeIndexEq = storeIndex
for i in range(storeIndex, right):
if l[i] == pivotValue:
l[storeIndexEq], l[i] = l[i], l[storeIndexEq]
storeIndexEq += 1
l[right], l[storeIndexEq] = l[storeIndexEq], l[right]
if n < storeIndex: return storeIndex
if n <= storeIndexEq: return n
return storeIndexEq
def medianofmedians(l, left, right, n):
while True:
if left == right: return left
pivotIndex = pivot(l, left, right)
pivotIndex = partition(l, left, right, pivotIndex, n)
if n == pivotIndex: return n
elif n < pivotIndex: right = pivotIndex - 1
else: left = pivotIndex + 1
def bfs(g, s):
n, L = len(g), {}
#for i in range(1, n+1): L[i] = 0
L[s] = None; S = [s]
while len(S) != 0:
u = S.pop(0)
for v in g[u]:
if not v in L: L[v] = u; S.append(v)
return L
def mbst(g, costs):
if len(g) == 2 and len(g[next(iter(g))]) == 1: return g
l = [(costs[(x, y)], (x, y)) for x in g for y in g[x] if x < y]
medianofmedians(l, 0, len(l) - 1, len(l) // 2)
A, B = l[len(l)//2:], l[:len(l)//2]
Gb = {}
for _, (x, y) in B:
if x > y: continue
if not x in Gb: Gb[x] = []
if not y in Gb: Gb[y] = []
Gb[x].append(y)
Gb[y].append(x)
F, Fr, S = {}, {}, set(Gb)
while len(S) != 0:
C = set(bfs(Gb, next(iter(S))))
S -= C
root = next(iter(C))
F[root] = C
for x in C: Fr[x] = root
if len(F) == 1 and len(Fr) == len(g): return mbst(Gb, costs)
else:
Ga, ca, mp = {}, {}, {}
for _, (x, y) in A:
if x > y or (x in Fr and y in Fr): continue
if x in Fr:
skip = (Fr[x], y) in ca
if not skip or ca[(Fr[x], y)] > costs[(x, y)]:
ca[(Fr[x], y)] = ca[(y, Fr[x])] = costs[(x, y)]
if skip: continue
mp[(Fr[x], y)] = (x, y); mp[(y, Fr[x])] = (y, x)
x = Fr[x]
elif y in Fr:
skip = (x, Fr[y]) in ca
if not skip or ca[(x, Fr[y])] > costs[(x, y)]:
ca[(x, Fr[y])] = ca[(Fr[y], x)] = costs[(x, y)]
if skip: continue
mp[(x, Fr[y])] = (x, y); mp[(Fr[y], x)] = (y, x)
y = Fr[y]
else: ca[(x, y)] = ca[(y, x)] = costs[(x, y)]
if not x in Ga: Ga[x] = []
if not y in Ga: Ga[y] = []
Ga[x].append(y)
Ga[y].append(x)
res = mbst(Ga, ca)
finres = {}
for x in res:
for y in res[x]:
if x > y: continue
if (x, y) in mp: x, y = mp[(x, y)]
if not x in finres: finres[x] = []
if not y in finres: finres[y] = []
finres[x].append(y)
finres[y].append(x)
for x in Gb:
for y in Gb[x]:
if x > y: continue
if not x in finres: finres[x] = []
if not y in finres: finres[y] = []
finres[x].append(y)
finres[y].append(x)
return finres
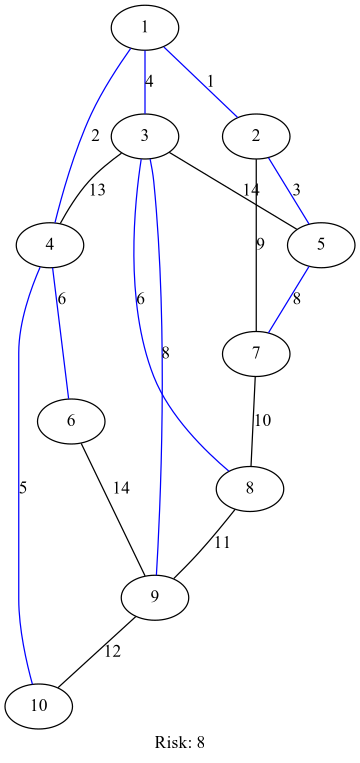
def camerini(adjmat):
g = mbst(adj_mat_to_edge_list(riskadjmat), adj_mat_to_costs(riskadjmat))
return {x-1: y-1 if not y is None else y for x, y in bfs(g, next(iter(g))).items()}
def operation_risk(riskadjmat):
mst = camerini(riskadjmat)
return max(riskadjmat[mst[x]][x] for x in mst if not mst[x] is None), mst
def risk_graph(adjmat, sr):
risk, spantree = sr
return ("graph {" +
"label=\"Risk: " + str(risk) + "\"" +
";".join(str(i+1) + "--" + str(j+1) + "[label=" + str(col) +
(";color=blue" if spantree[i] == j or spantree[j] == i else "") + "]"
for i, row in enumerate(adjmat) for j, col in enumerate(row) if i < j and not col is None) + "}")
riskadjmat = [[None, 1, 4, 2, None, None, None, None, None, None],
[1, None, None, None, 3, None, 9, None, None, None],
[4, None, None, 13, 14, None, None, 6, 8, None],
[2, None, 13, None, None, 6, None, None, None, 5],
[None, 3, 14, None, None, None, 8, None, None, None],
[None, None, None, 6, None, None, None, None, 14, None],
[None, 9, None, None, 8, None, None, 10, None, None],
[None, None, 6, None, None, None, 10, None, 11, None],
[None, None, 8, None, None, 14, None, 11, None, 12],
[None, None, None, 5, None, None, None, None, 12, None]]
import graphviz
graphviz.Source(risk_graph(riskadjmat, operation_risk(riskadjmat)))
![Wikipedia MBST example]()