I am always interested in the quickest ways to iterate over rows so I ran a few benchmarks of the other two answers and two more approaches. One is exactly the same as the answer by margusl except it uses purrr::pmap() rather than dplyr::rowwise(), which can be a faster alternative.
I also added an approach which converts to matrix, transposes and iterates over columns:
x <- t(as.matrix(df[, c("A", "B", "C", "D")]))
df$Z <- sapply(seq_len(nrow(df)), \(i){
overlaps <- intersect(
x[, i],
strsplit(df$X[[i]], ",")[[1]]
)
if (length(overlaps) == 0) NA_character_ else overlaps
})
This approach turns out to be much faster than the others. I also tried iterating over columns using data.table::transpose() and in this case it is almost as fast as t(), but not faster. So the message is, if you can, iterate over columns instead of rows.
Full benchmark results
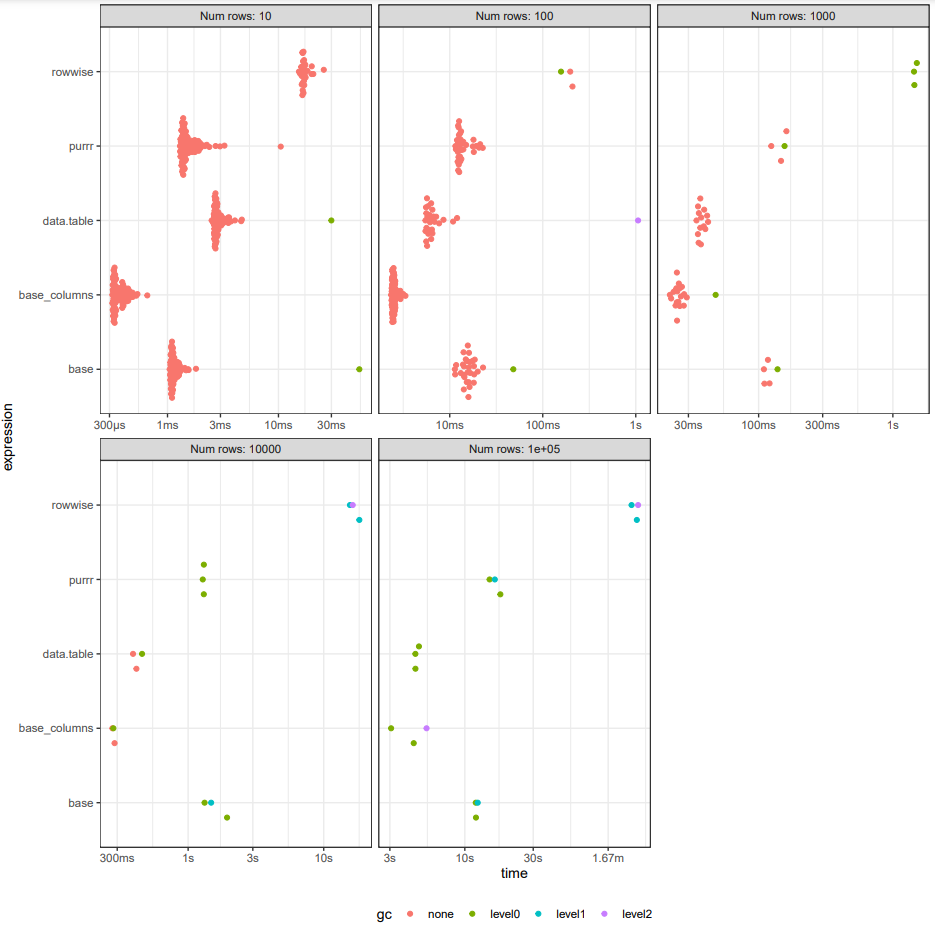
I generated random strings in the same format as your data frame for between 10 and 10^5 rows and compared the approaches. purrr::pmap() is comparable in speed to the base R approach. rowwise() is particularly slow with a lot more memory usage (500mb with 1e5 rows, compared with 800kb for Friede's base R approach). With 1e5 rows, the base R approach takes 13 seconds, purrr::pmap() 15 seconds, dplyr::rowwise() 2m40 seconds and iterating over columns takes 4 seconds. The full results are:
# A tibble: 25 × 10
expression n min median `itr/sec` mem_alloc `gc/sec` n_itr n_gc total_time
<bch:expr> <dbl> <dbl> <dbl> <dbl> <bch:byt> <dbl> <int> <dbl> <dbl>
1 base 10 1.05 1.12 860. 0B 8.69 99 1 115.
2 rowwise 10 15.3 16.8 57.6 53.25KB 0 29 0 503.
3 purrr 10 1.29 1.44 599. 10.57KB 0 100 0 167.
4 data.table 10 2.50 2.77 345. 187.04KB 3.48 99 1 287.
5 base_columns 10 0.319 0.347 2655. 736B 0 100 0 37.7
6 base 100 11.4 15.6 63.6 848B 2.19 29 1 456.
7 rowwise 100 198. 203. 4.92 510.5KB 2.46 2 1 406.
8 purrr 100 11.5 12.9 70.8 105.04KB 0 36 0 509.
9 data.table 100 5.48 6.14 151. 207.79KB 5.60 27 1 179.
10 base_columns 100 2.34 2.50 392. 9.09KB 0 100 0 255.
11 base 1000 110. 117. 8.38 7.86KB 1.68 5 1 596.
12 rowwise 1000 1437. 1447. 0.683 4.93MB 1.14 3 5 4393.
13 purrr 1000 124. 152. 6.80 1.02MB 1.70 4 1 588.
14 data.table 1000 34.5 37.0 26.6 381.34KB 0 14 0 527.
15 base_columns 1000 21.8 25.3 37.8 86.41KB 1.99 19 1 503.
16 base 10000 1322. 1479. 0.633 83.5KB 1.69 3 8 4738.
17 rowwise 10000 15603. 16366. 0.0597 49.18MB 1.19 3 60 50284.
18 purrr 10000 1282. 1306. 0.770 10.22MB 1.28 3 5 3894.
19 data.table 10000 392. 416. 2.37 2.12MB 0.790 3 1 1266.
20 base_columns 10000 276. 281. 3.55 909.53KB 1.18 3 1 844.
21 base 100000 11919. 11959. 0.0829 781.78KB 0.608 3 22 36184.
22 rowwise 100000 145657. 158549. 0.00644 491.72MB 0.403 3 188 465947.
23 purrr 100000 14887. 16189. 0.0615 102.23MB 0.390 3 19 48778.
24 data.table 100000 4521. 4526. 0.217 19.3MB 0.434 3 6 13832.
25 base_columns 100000 3061. 4401. 0.233 8.63MB 0.311 3 4 12870.
![enter image description here]()
Benchmark code
library(dplyr)
library(stringr)
library(data.table)
results <- bench::press(
n = 10^(1:5),
{
rand_strings <- CJ(letter = LETTERS[1:4], number = 0:9)[
,
strings := paste0(letter, number)
][]
df <- data.frame(
ID = seq(n),
A = sample(rand_strings[letter == "A", strings], n, replace = TRUE),
B = sample(rand_strings[letter == "B", strings], n, replace = TRUE),
C = sample(rand_strings[letter == "C", strings], n, replace = TRUE),
D = sample(rand_strings[letter == "D", strings], n, replace = TRUE),
X = sapply(
seq(n),
\(i)
paste(sample(rand_strings$strings, sample(0:10, 1), replace = TRUE), collapse = ",")
)
)
dt <- as.data.table(df)
bench::mark(
max_iterations = 100,
min_iterations = 3,
time_unit = "ms",
check = FALSE,
base = { # Friede approach
lapply(seq_len(nrow(df)), \(i) {
x <- df[i, LETTERS[1L:4L]]
x <- x[x %in% unlist(strsplit(df$X[i], ","))]
if (ncol(x)) paste0(x, collapse = ",") else NA
})
},
rowwise = { # margusl approach
df |>
rowwise() |>
mutate(Z = intersect(
c(across(A:D)),
str_split(X, ",", simplify = TRUE)
) |> str_c(collapse = ",") |> na_if("")) |>
ungroup()
},
purrr = {
df |>
purrr::pmap(\(ID, A, B, C, D, X) {
intersect(
c(A, B, C, D),
str_split(X, ",", simplify = TRUE)
) |>
str_c(collapse = ",") |>
na_if("")
}) |>
unlist()
},
data.table = {
x <- transpose(dt[, .(A, B, C, D)])
dt[, Z := sapply(seq_len(nrow(df)), \(i)
intersect(
x[[i]],
strsplit(df$X[[i]], ",")[[1]]
) |> paste(collapse = ","))][Z == "", Z := NA_character_]
}
)
}
)
# Plotting the results
library(ggplot2)
autoplot(results) +
facet_wrap(
vars(n),
scales = "free_x",
labeller = labeller(n = \(x) sprintf("Num rows: %s", x))
) +
theme_bw() +
theme(legend.position = "bottom")

rand_stringsis missing. Can you please add it in? – Heifetz