Looks like its possible to append row groups to already existing parquet file using fastparquet. This is quite a unique feature, since most libraries don't have this implementation.
Below is from pandas doc:
DataFrame.to_parquet(path, engine='auto', compression='snappy', index=None, partition_cols=None, **kwargs)
we have to pass in both engine and **kwargs.
- engine{‘auto’, ‘pyarrow’, ‘fastparquet’}
- **kwargs - Additional arguments passed to the parquet library.
**kwargs - here we need to pass is: append=True (from fastparquet)
import pandas as pd
from pathlib import Path
df = pd.DataFrame({'col1': [1, 2,], 'col2': [3, 4]})
file_path = Path("D:\\dev\\output.parquet")
if file_path.exists():
df.to_parquet(file_path, engine='fastparquet', append=True)
else:
df.to_parquet(file_path, engine='fastparquet')
If append is set to True and the file does not exist then you will see below error
AttributeError: 'ParquetFile' object has no attribute 'fmd'
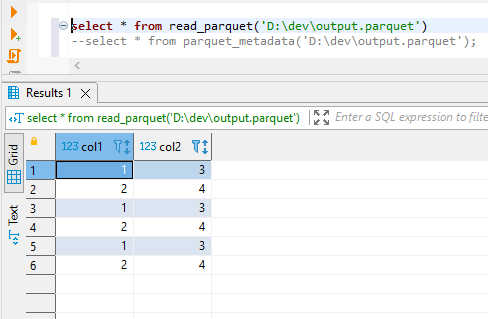
Running above script 3 times I have below data in parquet file.
![enter image description here]()
If I inspect the metadata, I can see that this resulted in 3 row groups.
![enter image description here]()
Note:
Append could be inefficient if you write too many small row groups. Typically recommended size of a row group is closer to 100,000 or 1,000,000 rows. This has a few benefits over very small row groups. Compression will work better, since compression operates within a row group only. There will also be less overhead spent on storing statistics, since each row group stores its own statistics.


appendmode forto_parquet()API.If you want to append to a file, theappendmode is for the file.That's what I try to express earlier. – Sonja