fitter provides an iteration process over possible fitting distributions.
It also outputs a plot and summary table with statistic values.
fitter package provides a simple class to identify the distribution
from which a data samples is generated from. It uses 80 distributions
from Scipy and allows you to plot the results to check what is the
most probable distribution and the best parameters.
So basically the same iterative fit test procedure as described in other answers, but conveniently executed by the module.
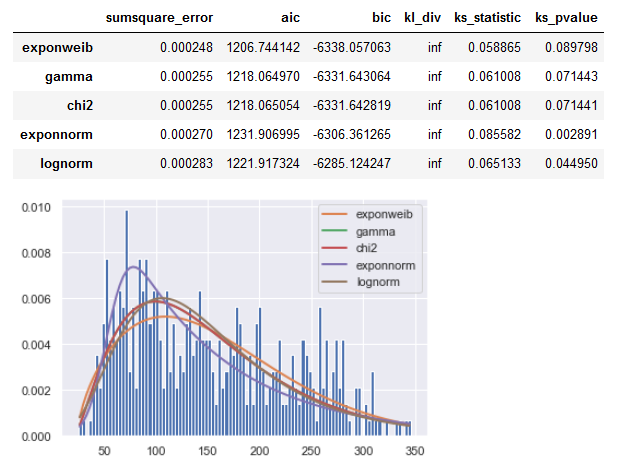
Result for your SR_y series:
![enter image description here]()
Code:
from sklearn.datasets import load_diabetes
from fitter import Fitter, get_common_distributions
#Get Data - from question
data = load_diabetes()
X, y_ = data.data, data.target
#Organize Data - from question
SR_y = pd.Series(y_, name="y_ (Target Vector Distribution)")
# fitter
distributions_set = get_common_distributions()
distributions_set.extend(['arcsine', 'cosine', 'expon', 'weibull_max', 'weibull_min',
'dweibull', 't', 'pareto', 'exponnorm', 'lognorm',
"norm", "exponweib", "weibull_max", "weibull_min", "pareto", "genextreme"])
f = Fitter(SR_y, distributions = distributions_set)
f.fit()
f.summary()
Parameters of those fitted distributions as a dict:
f.fitted_param
{'expon': (25.0, 127.13348416289594),
'cauchy': (132.95536663886972, 52.62243313109789),
'gamma': (2.496376511103246, 20.737715299081657, 52.63462302106953),
'norm': (152.13348416289594, 77.00574586945044),
'chi2': (4.9927545799818525, 20.737731375230684, 26.317289176495912),
'rayleigh': (14.700761411215545, 111.3948791009951),
'uniform': (25.0, 321.0),
'powerlaw': (1.0864390359784966, -6.82376066691087, 352.82376073752073),
'cosine': (159.01669793410446, 65.6033963343604),
'arcsine': (-6.99037533558757, 352.9903753355876),
'exponpow': (0.15440493125261756, 24.999999999999996, 16.00571403929016),
'weibull_max': (0.168196678837625, 346.0000000000001, 1.6686318895897978),
'weibull_min': (0.2750237375428041, 24.999999999999996, 6.998090013988461),
'dweibull': (1.6343449438402855, 157.0247145542748, 73.64165822064473),
'pareto': (0.6022461735477798, -0.06169932009129858, 25.06169863339018),
'exponnorm': (6.298770105099791, 53.6065309642624, 15.642251691931591),
't': (127967.50529392948, 152.12481045573628, 76.98521783304597),
'exponweib': (0.9662752277542657, 1.6900600238468133, 24.142487003378918, 150.25955880342326),
'lognorm': (0.44469088248930166, -29.00650970868123, 164.71283014005542),
'genextreme': (0.029317901766728702, 116.52312667345038, 63.454691756821106)}
To get a list of all available distributions:
from fitter import get_distributions
get_distributions()
Testing for all of them takes a long time, so it's best to use the implemened get_common_distributions() and potentially extend them with likely distribution as done in the code above.
Make sure to have the current (1.4.1 or newer) fitter version installed:
import fitter
print(fitter.version)
I had logging errors with a previous one and for my conda environment I needed:
conda install -c bioconda fitter


scipy.statsdistributions available, perhaps you can combine a few of these to generate your desired distribution. – Kynan