I get (working from the shared file is a good idea on this case):
Loading with:
data = io.loadmat('../Downloads/anno_bbox.mat')
I get:
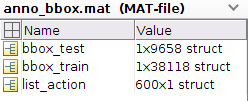
In [96]: data['bbox_test'].dtype
Out[96]: dtype([('filename', 'O'), ('size', 'O'), ('hoi', 'O')])
In [97]: data['bbox_test'].shape
Out[97]: (1, 9658)
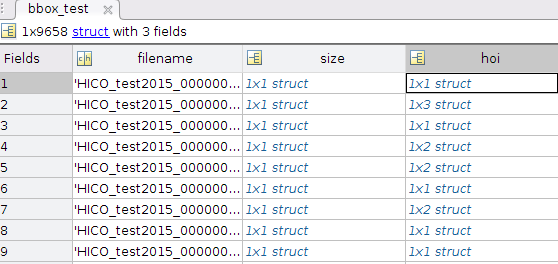
I could have assigned bbox_test=data['bbox_test']. This variable has 9658 records, with three fields, each with object dtype.
So there's a filename (a string embedded in a 1 element array)
In [101]: data['bbox_test'][0,0]['filename']
Out[101]: array(['HICO_test2015_00000001.jpg'], dtype='<U26')
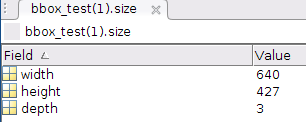
size has 3 fields, with 3 numbers embedded in arrays (2d matlab matrices):
In [102]: data['bbox_test'][0,0]['size']
Out[102]:
array([[(array([[640]], dtype=uint16), array([[427]], dtype=uint16), array([[3]], dtype=uint8))]],
dtype=[('width', 'O'), ('height', 'O'), ('depth', 'O')])
In [112]: data['bbox_test'][0,0]['size'][0,0].item()
Out[112]:
(array([[640]], dtype=uint16),
array([[427]], dtype=uint16),
array([[3]], dtype=uint8))
hoi is more complicated:
In [103]: data['bbox_test'][0,0]['hoi']
Out[103]:
array([[(array([[246]], dtype=uint8), array([[(array([[320]], dtype=uint16), array([[359]], dtype=uint16), array([[306]], dtype=uint16), array([[349]], dtype=uint16)),...
dtype=[('id', 'O'), ('bboxhuman', 'O'), ('bboxobject', 'O'), ('connection', 'O'), ('invis', 'O')])
In [126]: data['bbox_test'][0,1]['hoi']['id']
Out[126]:
array([[array([[132]], dtype=uint8), array([[140]], dtype=uint8),
array([[144]], dtype=uint8)]], dtype=object)
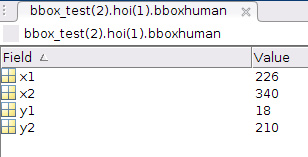
In [130]: data['bbox_test'][0,1]['hoi']['bboxhuman'][0,0]
Out[130]:
array([[(array([[226]], dtype=uint8), array([[340]], dtype=uint16), array([[18]], dtype=uint8), array([[210]], dtype=uint8))]],
dtype=[('x1', 'O'), ('x2', 'O'), ('y1', 'O'), ('y2', 'O')])
So the data that you show in the MATLAB structures is all there, in a nested structure of arrays (often 2d (1,1) shape), object dtype or multiple fields.
Going back and loading with squeeze_me I get a simpler:
In [133]: data['bbox_test'][1]['hoi']['bboxhuman']
Out[133]:
array([array((226, 340, 18, 210),
dtype=[('x1', 'O'), ('x2', 'O'), ('y1', 'O'), ('y2', 'O')]),
array((230, 356, 19, 212),
dtype=[('x1', 'O'), ('x2', 'O'), ('y1', 'O'), ('y2', 'O')]),
array((234, 342, 13, 202),
dtype=[('x1', 'O'), ('x2', 'O'), ('y1', 'O'), ('y2', 'O')])],
dtype=object)
With struct_as_record='False', I get
In [136]: data['bbox_test'][1]
Out[136]: <scipy.io.matlab.mio5_params.mat_struct at 0x7f90841e9748>
Looking at the attributes of this rec I see I can access 'fields' by attribute name:
In [137]: rec = data['bbox_test'][1]
In [138]: rec.filename
Out[138]: 'HICO_test2015_00000002.jpg'
In [139]: rec.size
Out[139]: <scipy.io.matlab.mio5_params.mat_struct at 0x7f90841e9b38>
In [141]: rec.size.width
Out[141]: 640
In [142]: rec.hoi
Out[142]:
array([<scipy.io.matlab.mio5_params.mat_struct object at 0x7f90841e9be0>,
<scipy.io.matlab.mio5_params.mat_struct object at 0x7f90841e9e10>,
<scipy.io.matlab.mio5_params.mat_struct object at 0x7f90841ee0b8>],
dtype=object)
In [145]: rec.hoi[1].bboxhuman
Out[145]: <scipy.io.matlab.mio5_params.mat_struct at 0x7f90841e9f98>
In [146]: rec.hoi[1].bboxhuman.x1
Out[146]: 230
In [147]: vars(rec.hoi[1].bboxhuman)
Out[147]:
{'_fieldnames': ['x1', 'x2', 'y1', 'y2'],
'x1': 230,
'x2': 356,
'y1': 19,
'y2': 212}
and so on.