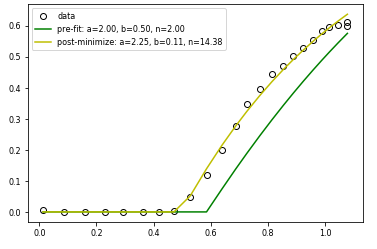
I am trying to fit the functiony= 1-a(1-bx)**n to some experimental data using scipy curve_fit. The model only exists for y>0, so I clip the calculated values to enforce this.
The code is shown below
import numpy as np
import scipy.optimize
import matplotlib.pyplot as plt
# Driver function for scipy.minimize
def driver_func(x, xobs, yobs):
# Evaluate the fit function with the current parameter estimates
ynew = myfunc(xobs, *x)
yerr = np.sum((ynew - yobs) ** 2)
return yerr
# Define function
def myfunc(x, a, b, n):
y = 1.0 - a * np.power(1.0 - b * x, n)
y = np.clip(y, 0.00, None )
return y
if __name__ == "__main__":
# Initialise data
yobs = np.array([0.005, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.004,
0.048, 0.119, 0.199, 0.277, 0.346, 0.395, 0.444, 0.469,
0.502, 0.527, 0.553, 0.582, 0.595, 0.603, 0.612, 0.599])
xobs = np.array([0.013, 0.088, 0.159, 0.230, 0.292, 0.362, 0.419, 0.471,
0.528, 0.585, 0.639, 0.687, 0.726, 0.772, 0.814, 0.854,
0.889, 0.924, 0.958, 0.989, 1.015, 1.045, 1.076, 1.078])
# Initial guess
p0 = [2.0, 0.5, 2.0]
# Check fit pre-regression
yold = myfunc(xobs, *p0)
plt.plot(xobs, yobs, 'ko', label='data', fillstyle='none')
plt.plot(xobs, yold, 'g-', label='pre-fit: a=%4.2f, b=%4.2f, n=%4.2f' % tuple(p0))
# Fit curve using SCIPY CURVE_FIT
try:
popt, pcov = scipy.optimize.curve_fit(myfunc, xobs, yobs, p0=p0)
except:
print("Could not fit data using SCIPY curve_fit")
else:
ynew = myfunc(xobs, *popt)
plt.plot(xobs, ynew, 'r-', label='post-curve_fit: a=%4.2f, b=%4.2f, n=%4.2f' % tuple(popt))
# Fit curve using SCIPY MINIMIZE
res = scipy.optimize.minimize(driver_func, p0, args=(xobs, yobs), method='Nelder-Mead')
ynw2 = myfunc(xobs, *res.x)
plt.plot(xobs, ynw2, 'y-', label='post-minimize: a=%4.2f, b=%4.2f, n=%4.2f' % tuple(res.x))
plt.legend()
plt.show()
I also used SCIPY MINIMIZE to achieve the same thing. As the image below shows, MINIMIZE works, but CURVE_FIT basically runs out of evalautions and gives up, even though the starting guess is not that far away from the MINIMIZE solution (at least visually). Would appreciate any thoughts on why curve_fit seems not to be working here.
Thanks!
Update: Following the comments by mikuszefski I made the following adjustments 1. removed the clipping from the fit function as follows:
def myfunc_noclip(x, a, b, n):
y = 1.0 - a * np.power(1.0 - b * x, n)
return y
introduced clipped arrays by removing data below a threshold
ymin = 0.01 xclp = xobs[np.where(yobs >= ymin)] yclp = yobs[np.where(yobs >= ymin)]improved the initial guess (again visually)
p0 = [1.75, 0.5, 2.0]updated the call to curve_fit
popt, pcov = scipy.optimize.curve_fit(myfunc_noclip, xclp, yclp, p0=p0)
But this does not seem to have helped as the following plot shows:
Other posts on stackoverflow seem to suggest that scipy curve_fit has trouble fitting curves where one of the fit parameters is an exponent eg
SciPy curve_fit not working when one of the parameters to fit is a power
so I am guessing that I have the same problem. Not sure how to solve it though ...