Forewords
I came across this problem this week and the only relevant issue I have found about it is this post. I have almost same requirement as the OP:
But I also need:
- To be able to perform statistical test as well;
- While being compliant with the
scipy random variable interface.
As @Jacques Gaudin pointed out the interface for rv_continous (see distribution architecture for details) does not ensure follow up for loc and scale parameters when inheriting from this class. And this is somehow misleading and unfortunate.
Implementing the __init__ method of course allow to create the missing binding but the trade off is: it breaks the pattern scipy is currently using to implement random variables (see an example of implementation for lognormal).
So, I took time to dig into the scipy code and I have created a MCVE for this distribution. Although it is not totally complete (it mainly misses moments overrides) it fits the bill for both OP and my purposes while having satisfying accuracy and performance.
MCVE
An interface compliant implementation of this random variable could be:
class logitnorm_gen(stats.rv_continuous):
def _argcheck(self, m, s):
return (s > 0.) & (m > -np.inf)
def _pdf(self, x, m, s):
return stats.norm(loc=m, scale=s).pdf(special.logit(x))/(x*(1-x))
def _cdf(self, x, m, s):
return stats.norm(loc=m, scale=s).cdf(special.logit(x))
def _rvs(self, m, s, size=None, random_state=None):
return special.expit(m + s*random_state.standard_normal(size))
def fit(self, data, **kwargs):
return stats.norm.fit(special.logit(data), **kwargs)
logitnorm = logitnorm_gen(a=0.0, b=1.0, name="logitnorm")
This implementation unlock most of the scipy random variables potential.
N = 1000
law = logitnorm(0.24, 1.31) # Defining a RV
sample = law.rvs(size=N) # Sampling from RV
params = logitnorm.fit(sample) # Infer parameters w/ MLE
check = stats.kstest(sample, law.cdf) # Hypothesis testing
bins = np.arange(0.0, 1.1, 0.1) # Bin boundaries
expected = np.diff(law.cdf(bins)) # Expected bin counts
As it relies on scipy normal distribution we may assume underlying functions have the same accuracy and performance than normal random variable object. But it might indeed be subject to float arithmetic inaccuracy especially when dealing with highly skewed distributions at the support boundary.
Tests
To check out how it performs we draw some distribution of interest and check them.
Let's create some fixtures:
def generate_fixtures(
locs=[-2.0, -1.0, 0.0, 0.5, 1.0, 2.0],
scales=[0.32, 0.56, 1.00, 1.78, 3.16],
sizes=[100, 1000, 10000],
seeds=[789, 123456, 999999]
):
for (loc, scale, size, seed) in itertools.product(locs, scales, sizes, seeds):
yield {"parameters": {"loc": loc, "scale": scale}, "size": size, "random_state": seed}
And perform checks on related distributions and samples:
eps = 1e-8
x = np.linspace(0. + eps, 1. - eps, 10000)
for fixture in generate_fixtures():
# Reference:
parameters = fixture.pop("parameters")
normal = stats.norm(**parameters)
sample = special.expit(normal.rvs(**fixture))
# Logit Normal Law:
law = logitnorm(m=parameters["loc"], s=parameters["scale"])
check = law.rvs(**fixture)
# Fit:
p = logitnorm.fit(sample)
trial = logitnorm(*p)
resample = trial.rvs(**fixture)
# Hypothetis Tests:
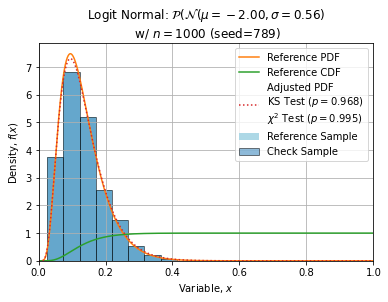
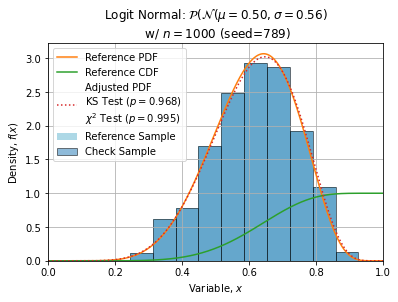
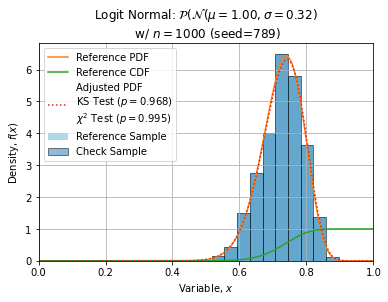
ks = stats.kstest(check, trial.cdf)
bins = np.histogram(resample)[1]
obs = np.diff(trial.cdf(bins))*fixture["size"]
ref = np.diff(law.cdf(bins))*fixture["size"]
chi2 = stats.chisquare(obs, ref, ddof=2)
Some adjustments with n=1000, seed=789 (this sample is quite normal) are shown below:
![enter image description here]()
![enter image description here]()
![enter image description here]()
![enter image description here]()


LogitNormal().pdf? I guess you should callLogitNormal()._pdf. Cause_pdfis how you called the only method inLogitNormalclass. – Aurita_pdfor_cdfis how you subclassrv_continuousaccording to the documentation docs.scipy.org/doc/scipy/reference/generated/… and also the post that I linked in my question. – Propoundrv_continuous.pdfmethod performs transformation ofxvalues usinglocandscaleparameters. I guess you will get the same result with defaults:sigma, mu = 1, 0. – Auritarv_continuous.pdfcomputes probability density function. Andscipy.stats.norm.pdfcomputes probability density function. I guess that's all I can tell since I'm not familiar with these functions. Wish you luck. – Aurita