I am trying to obtain a double Gaussian distribution for data (link) using Python. The raw data is of the form:
For the given data, I would like to obtain two Gaussian profiles for the peaks seen in figure. I tried it with the following code (source):
from sklearn import mixture
import matplotlib.pyplot
import matplotlib.mlab
import numpy as np
from pylab import *
data = np.genfromtxt('gaussian_fit.dat', skiprows = 1)
x = data[:, 0]
y = data[:, 1]
clf = mixture.GMM(n_components=2, covariance_type='full')
clf.fit((y, x))
m1, m2 = clf.means_
w1, w2 = clf.weights_
c1, c2 = clf.covars_
fig = plt.figure(figsize = (5, 5))
plt.subplot(111)
plotgauss1 = lambda x: plot(x,w1*matplotlib.mlab.normpdf(x,m1,np.sqrt(c1))[0], linewidth=3)
plotgauss2 = lambda x: plot(x,w2*matplotlib.mlab.normpdf(x,m2,np.sqrt(c2))[0], linewidth=3)
fig.savefig('gaussian_fit.pdf')
But I am not able to get the desired output. So, how can a double Gaussian distribution be obtained in Python?
Update
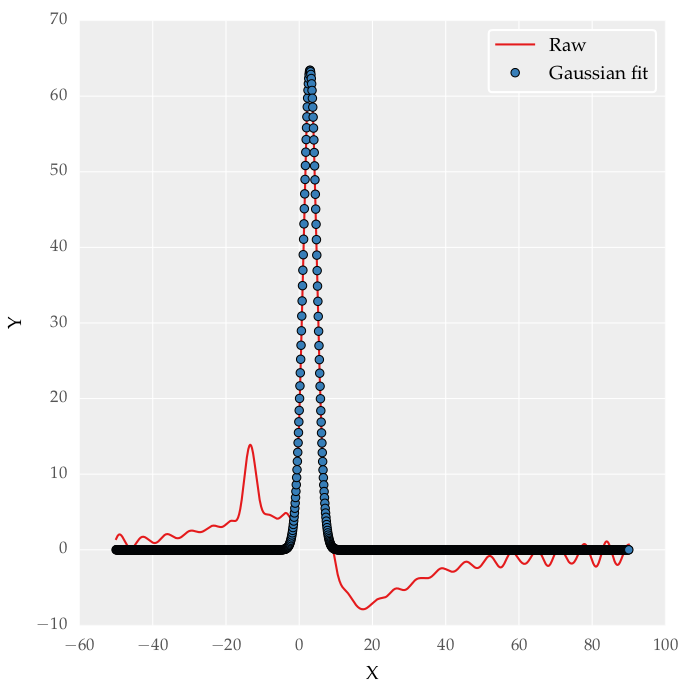
I was able to fit a single Gaussian distribution with the following code:
import pylab as plb
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
from scipy import asarray as ar,exp
import numpy as np
data = np.genfromtxt('gaussian_fit.dat', skiprows = 1)
x = data[:, 0]
y = data[:, 1]
n = len(x)
mean = sum(x*y)/n
sigma = sum(y*(x-mean)**2)/n
def gaus(x,a,x0,sigma):
return a*exp(-(x-x0)**2/(2*sigma**2))
popt,pcov = curve_fit(gaus, x, y ,p0 = [1, mean, sigma])
fig = plt.figure(figsize = (5, 5))
plt.subplot(111)
plt.plot(x, y, label='Raw')
plt.plot(x, gaus(x, *popt), 'o', markersize = 4, label='Gaussian fit')
plt.xlabel('X')
plt.ylabel('Y')
plt.legend()
fig.savefig('gaussian_fit.pdf')