In Python, suppose that I have continuous variables x and y, whose values are bounded between 0 and 1 (to make it easier). My assumption has always been that if I want to convert those variables into ordinal values with bins going like 0,0.01,0.02,...,0.98,0.99,1 one could simply round the original values to the second digit. For some reason, when I do that it leaves artifacts.
Let me illustrate the problem (notice however that my question is not how to get the correct plot, but actually how about doing the right binning). First these are the only modules one needs to reproduce the problem:
import numpy as np
import matplotlib.pyplot as plt
Now, suppose that we have continuous have data generated like the following (other data generating processes would also give the same issue):
# number of points drawn from Gaussian dists.:
n = 100000
x = np.random.normal(0, 2, n)
y = np.random.normal(4, 5, n)
# normalizing x and y to bound them between 0 and 1
# (it's way easier to illustrate the problem this way)
x = (x - min(x))/(max(x) - min(x))
y = (y - min(y))/(max(y) - min(y))
Then, let's convert x and y to ordinal in the above mentioned interval just by applying some rounding. Then, let's store results into a x by y matrix in order to plot its heatmap for illustration purposes:
# matrix that will represent the bins. Notice that the
# desired bins are every 0.01, from 0 to 1, so 100 bins:
mtx = np.zeros([100,100])
for i in range(n):
# my idea was that I could roughly get the bins by
# simply rounding to the 2nd decimal point:
posX = round(x[i], 2)
posY = round(y[i], 2)
mtx[int(posX*100)-1, int(posY*100)-1] += 1
I would expect the above to work, but when I plot the contents of the matrix mtx, I actually get weird artifacts. The code:
# notice, however, the weird close-to-empty lines at
# 0.30 and 0.59 of both x and y. This happens regardless
# of how I generate x and y. Regardless of distributions
# or of number of points (even if it obviously becomes
# impossible to see if there are too few points):
plt.matshow(mtx, cmap=plt.cm.jet)
plt.show(block=False)
Gives me:
The weirdest thing is that regardless of which distribution I use to generate x and y or which seed I use for the RNG, I always get the same horizontal and vertical near-empty lines at 0.30 and 0.59 of both x and y, quite often with the lines immediately parallel to those showing concentration of points (like you see in the image).
When I print value by value from that matrix to the console, I can actually confirm that the ones corresponding to those near-empty lines are indeed either zero or very close to zero - differently from their neighbor points.
My question can be more properly be divided into 2 pieces:
Why would the above happen? I genuinely would like to understand what exactly gives such a problem in that simple code.
What would be a better way to generate the
xbyymatrix that bins the values according to cutpoints 0,0.01,0.02,...,0.98,0.99,1 without leaving the artifacts above?
If one wants to easily grab the whole example code used above directly in one piece, here is the link: https://www.codepile.net/pile/VLAq4kLp
NOTE: I don't want to find a correct way of plotting. I want to find myeself the correct way of generating the "binned values matrix" that is represented is the above plot. I know that there are other ways to accomplish the heatmap plotting without the artifacts, for instance using plt.matshow(mtx, cmap=plt.cm.jet); plt.show(block=False) or plt.hist2d(x, y, bins=100). What I am asking is where is the problem in my matrix generation itself, which creates those near-zero elements.

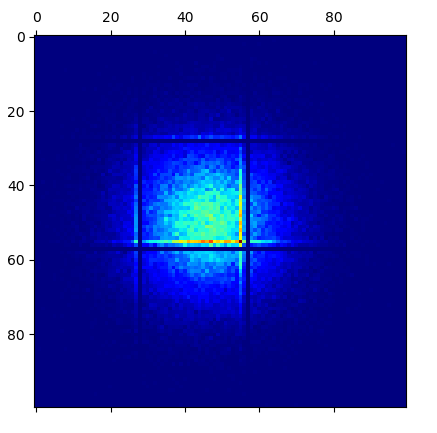
matplotlib? Reproduced your code here, don't see anything wrong with matrix elements. – Amphistylarnumpy.histogram2d? That would solve this problem in a single line and be much faster than a loop. – Predicatenp.savetxt("filename", mtx), then open the saved file with a spreadsheet software and check around rows/columns 30 and 59. It does not matter how many times I repeat the code, the problem is always there. – Disclaimnumpy.histogram2dsolves it (easy to check withplt.matshow(np.histogram2d(x,y,bins=100)[0]);plt.show()in my original code). But I am trying to precisely understand and solve what is the problem you identified in the algorithm (thta creates the mentioned problem) with which I was trying to (rather simplistically) discretize the continuous values usinground. – Disclaim