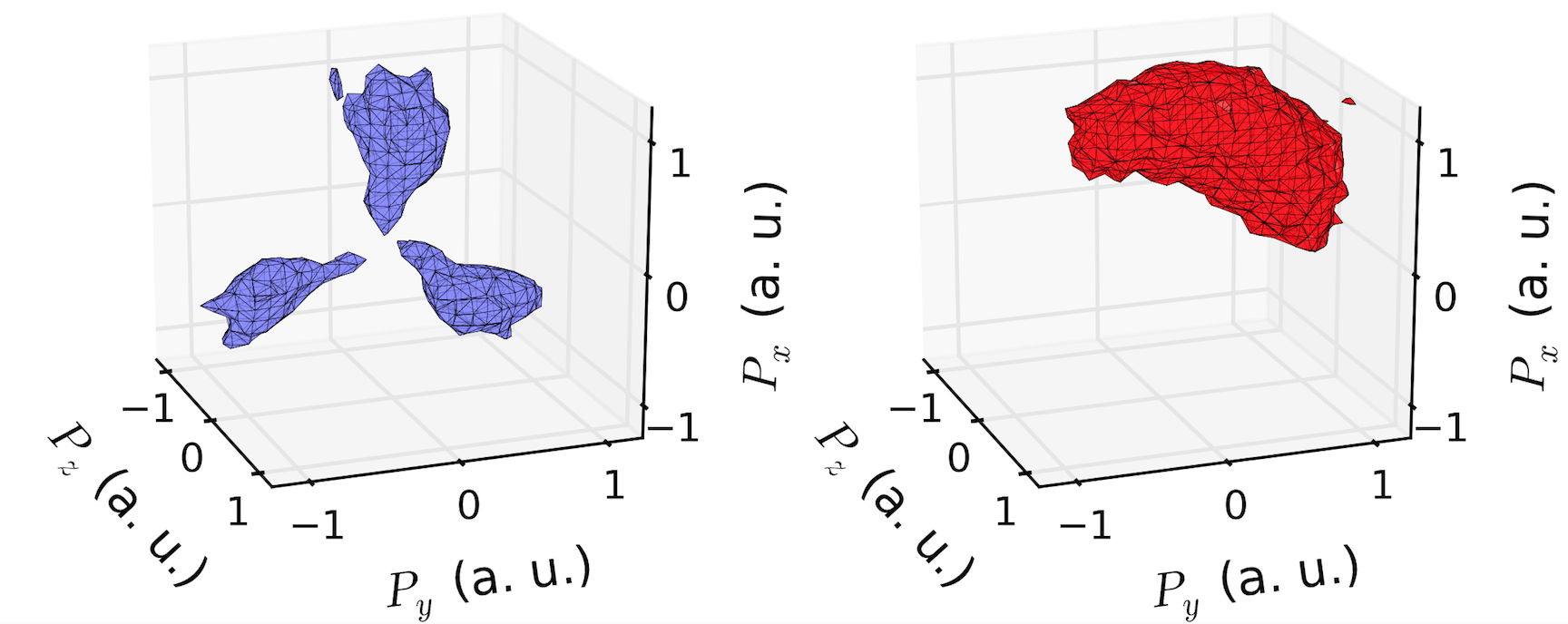
Complementing the answer of @DanHickstein, you can also use trisurf to visualize the polygons obtained in the marching cubes phase.
import numpy as np
from numpy import sin, cos, pi
from skimage import measure
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
def fun(x, y, z):
return cos(x) + cos(y) + cos(z)
x, y, z = pi*np.mgrid[-1:1:31j, -1:1:31j, -1:1:31j]
vol = fun(x, y, z)
iso_val=0.0
verts, faces = measure.marching_cubes(vol, iso_val, spacing=(0.1, 0.1, 0.1))
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
ax.plot_trisurf(verts[:, 0], verts[:,1], faces, verts[:, 2],
cmap='Spectral', lw=1)
plt.show()
![enter image description here]()
Update: May 11, 2018
As mentioned by @DrBwts, now marching_cubes return 4 values. The following code works.
import numpy as np
from numpy import sin, cos, pi
from skimage import measure
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
def fun(x, y, z):
return cos(x) + cos(y) + cos(z)
x, y, z = pi*np.mgrid[-1:1:31j, -1:1:31j, -1:1:31j]
vol = fun(x, y, z)
iso_val=0.0
verts, faces, _, _ = measure.marching_cubes(vol, iso_val, spacing=(0.1, 0.1, 0.1))
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
ax.plot_trisurf(verts[:, 0], verts[:,1], faces, verts[:, 2],
cmap='Spectral', lw=1)
plt.show()
Update: February 2, 2020
Adding to my previous answer, I should mention that since then PyVista has been released, and it makes this
kind of tasks somewhat effortless.
Following the same example as before.
from numpy import cos, pi, mgrid
import pyvista as pv
#%% Data
x, y, z = pi*mgrid[-1:1:31j, -1:1:31j, -1:1:31j]
vol = cos(x) + cos(y) + cos(z)
grid = pv.StructuredGrid(x, y, z)
grid["vol"] = vol.flatten()
contours = grid.contour([0])
#%% Visualization
pv.set_plot_theme('document')
p = pv.Plotter()
p.add_mesh(contours, scalars=contours.points[:, 2], show_scalar_bar=False)
p.show()
With the following result
![enter image description here]()
Update: February 24, 2020
As mentioned by @HenriMenke, marching_cubes has been renamed to marching_cubes_lewiner. The "new" snippet is the following.
import numpy as np
from numpy import cos, pi
from skimage.measure import marching_cubes_lewiner
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
x, y, z = pi*np.mgrid[-1:1:31j, -1:1:31j, -1:1:31j]
vol = cos(x) + cos(y) + cos(z)
iso_val=0.0
verts, faces, _, _ = marching_cubes_lewiner(vol, iso_val, spacing=(0.1, 0.1, 0.1))
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
ax.plot_trisurf(verts[:, 0], verts[:,1], faces, verts[:, 2], cmap='Spectral',
lw=1)
plt.show()